Usage
## S3 method for class 'rootogram'
plot(
x,
style = NULL,
scale = NULL,
expected = NULL,
ref = NULL,
confint = NULL,
confint_level = 0.95,
confint_type = c("tukey", "pointwise", "simultaneous"),
confint_nrep = 1000,
xlim = c(NA, NA),
ylim = c(NA, NA),
xlab = NULL,
ylab = NULL,
main = NULL,
axes = TRUE,
box = FALSE,
col = "darkgray",
border = "black",
lwd = 1,
lty = 1,
alpha_min = 0.8,
expected_col = 2,
expected_pch = 19,
expected_lty = 1,
expected_lwd = 2,
confint_col = "black",
confint_lty = 2,
confint_lwd = 1.75,
ref_col = "black",
ref_lty = 1,
ref_lwd = 1.25,
...
)
## S3 method for class 'rootogram'
autoplot(
object,
style = NULL,
scale = NULL,
expected = NULL,
ref = NULL,
confint = NULL,
confint_level = 0.95,
confint_type = c("tukey", "pointwise", "simultaneous"),
confint_nrep = 1000,
xlim = c(NA, NA),
ylim = c(NA, NA),
xlab = NULL,
ylab = NULL,
main = NULL,
legend = FALSE,
theme = NULL,
colour = "black",
fill = "darkgray",
size = 0.5,
linetype = 1,
alpha = NA,
expected_colour = 2,
expected_size = 1,
expected_linetype = 1,
expected_alpha = 1,
expected_fill = NA,
expected_stroke = 0.5,
expected_shape = 19,
confint_colour = "black",
confint_size = 0.5,
confint_linetype = 2,
confint_alpha = NA,
ref_colour = "black",
ref_size = 0.5,
ref_linetype = 1,
ref_alpha = NA,
...
)
Details
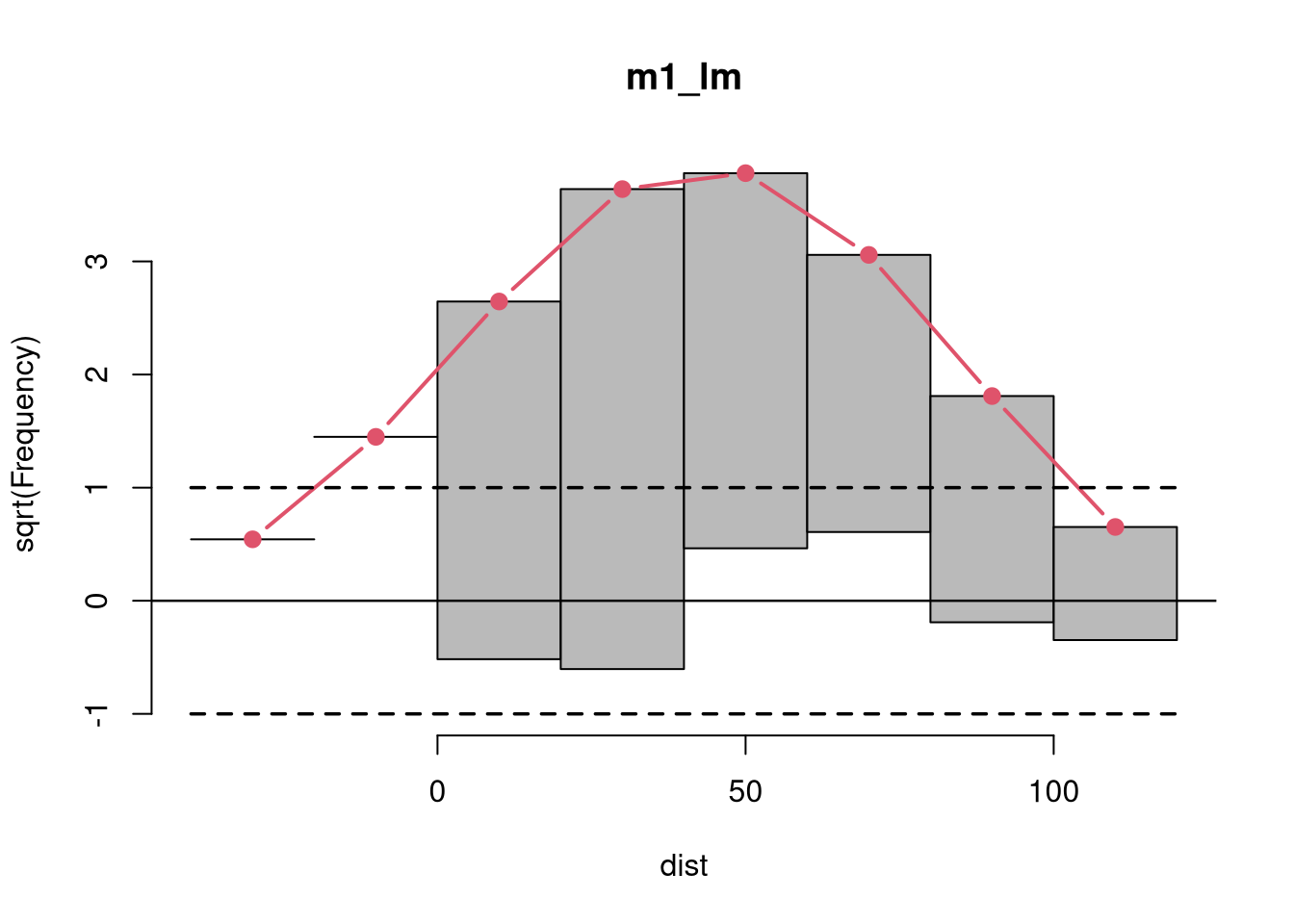
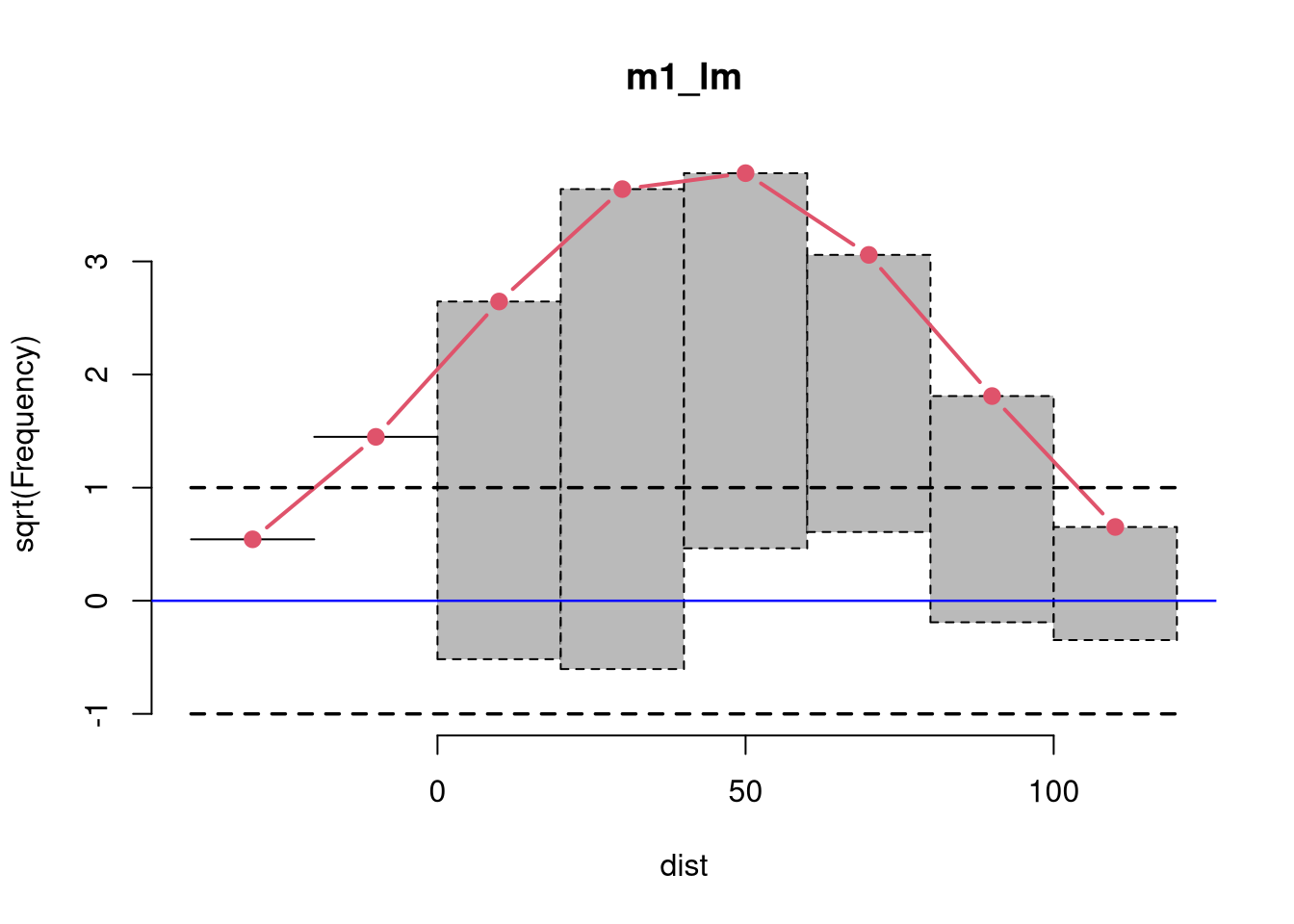
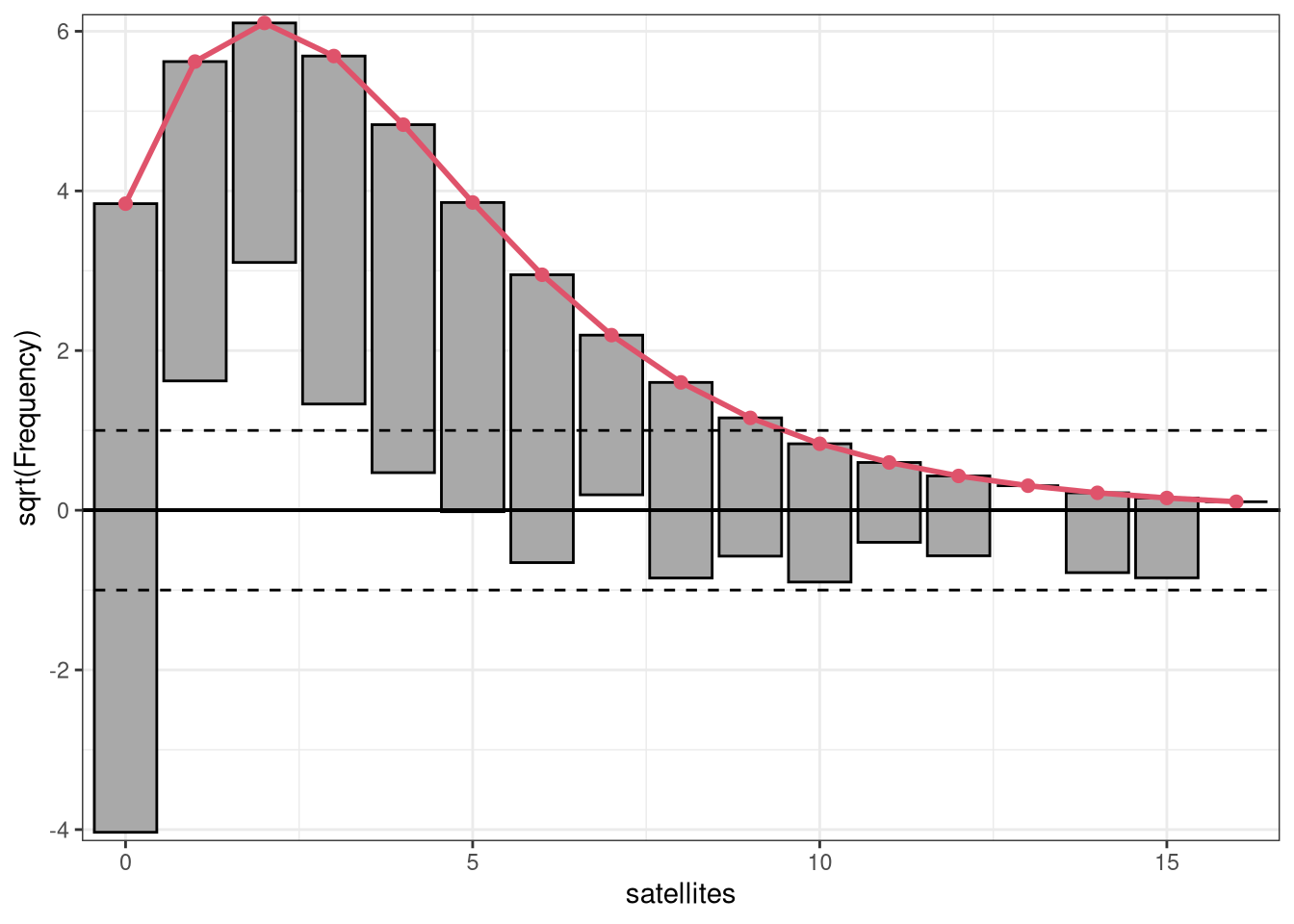
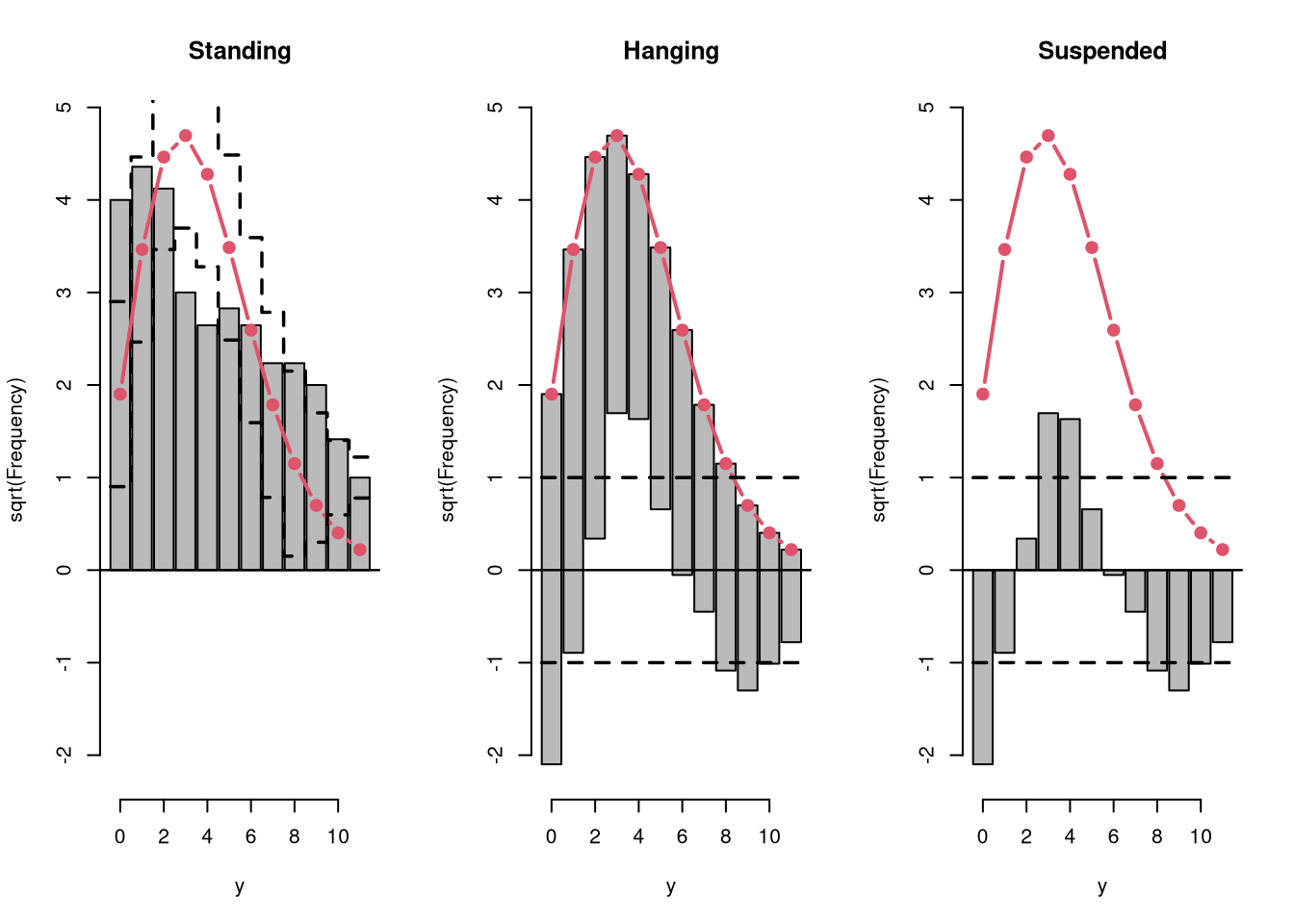
Rootograms graphically compare (square roots) of empirical frequencies with expected (fitted) frequencies from a probability model. For the observed distribution the histogram is drawn on a square root scale (hence the name) and superimposed with a line for the expected frequencies. The histogram can be “standing” on the x-axis (as usual), or “hanging” from the expected (fitted) curve, or a “suspended” histogram of deviations can be drawn.
Rootograms are associated with the work of John W. Tukey (see Tukey 1977) and were originally proposed for assessing the goodness of fit of univariate distributions and extended by Kleiber and Zeileis (2016) to regression setups.
As the expected distribution is typically a sum of different conditional distributions in regression models, the “pointwise” confidence intervals for each bin can be computed from mid-quantiles of a Poisson-Binomial distribution (Wilson and Einbeck 2021). Corresponding “simultaneous” confidence intervals for all bins can be obtained via simulation from the Poisson-Binomial distributions. As the pointwise confidence intervals are typically not substantially different from the warning limits of Tukey (1972, p. 61), set at +/- 1, these “tukey” intervals are used by default.
Note that for computing the exact “pointwise” intervals from the Poisson-Binomial distribution, the PoissonBinomial needs to be installed. Otherwise, a warning is issueed and a normal approximation is used.
References
Kleiber C, Zeileis A (2016). “Visualizing Count Data Regressions Using Rootograms.” The American Statistician, 70(3), 296–303. doi:10.1080/00031305.2016.1173590
Tukey JW (1972), “Some Graphic and Semigraphic Displays,” in Statistical Papers in Honor of George W. Snedecor, pp.293–316. Bancroft TA (Ed.). Iowa State University Press, Ames. Reprinted in William S. Cleveland (Ed.) (1988). The Collected Works of John W. Tukey, Volume V. Graphics: 1965–1985, Wadsworth & Brooks/Cole, Pacific Grove.
Tukey JW (1977). Exploratory Data Analysis. Addison-Wesley, Reading.
Wilson P, Einbeck J (2021). “A Graphical Tool for Assessing the Suitability of a Count Regression Model”, Austrian Journal of Statistics, 50(1), 1–23. doi:10.17713/ajs.v50i1.921