library("betareg")
## IGNORE_RDIFF_BEGIN
## package and data
library("betareg")
library("bamlss")
data("ReadingSkills", package = "betareg")
## classical beta regression via ML
rs1 <- betareg(accuracy ~ dyslexia * iq | dyslexia + iq, data = ReadingSkills)
## Bayesian additive model (with low number of iterations to speed up the example)
set.seed(0)
rs2 <- bamlss(accuracy ~ s(iq, by = dyslexia) | dyslexia + iq, data = ReadingSkills,
family = betar_family(), eps = 1e-7, n.iter = 400, burnin = 100)AICc -0.8829 logPost -64.6792 logLik 21.6241 edf 13.973 eps 1.0000 iteration 1
AICc -51.8915 logPost -25.8044 logLik 39.7035 edf 10.242 eps 2.8731 iteration 2
AICc -68.1251 logPost -7.3858 logLik 46.8183 edf 9.6642 eps 0.5561 iteration 3
AICc -73.1290 logPost 4.7269 logLik 48.3399 edf 9.0783 eps 0.0895 iteration 4
AICc -74.7842 logPost 5.5545 logLik 49.1675 edf 9.0783 eps 0.0629 iteration 5
AICc -75.7464 logPost 6.0357 logLik 49.6487 edf 9.0783 eps 0.0559 iteration 6
AICc -76.3072 logPost 6.3160 logLik 49.9291 edf 9.0783 eps 0.2490 iteration 7
AICc -77.3399 logPost 6.8324 logLik 50.4454 edf 9.0783 eps 0.2694 iteration 8
AICc -77.6557 logPost 6.9903 logLik 50.6033 edf 9.0783 eps 0.0207 iteration 9
AICc -77.8176 logPost 7.0713 logLik 50.6843 edf 9.0783 eps 0.0123 iteration 10
AICc -80.2391 logPost 8.7295 logLik 50.7321 edf 8.3559 eps 0.0099 iteration 11
AICc -81.1323 logPost -61.6299 logLik 50.7922 edf 8.1090 eps 0.0333 iteration 12
AICc -81.3164 logPost -654.389 logLik 50.8351 edf 8.0774 eps 0.0089 iteration 13
AICc -81.3726 logPost -4694.96 logLik 50.8579 edf 8.0740 eps 0.0054 iteration 14
AICc -81.4322 logPost -25766.3 logLik 50.8816 edf 8.0700 eps 0.0077 iteration 15
AICc -81.6598 logPost -2043.53 logLik 50.9498 edf 8.0406 eps 0.0179 iteration 16
AICc -82.3830 logPost -8852.97 logLik 51.1594 edf 7.9421 eps 0.0137 iteration 17
AICc -82.7904 logPost -13085.5 logLik 51.2898 edf 7.8944 eps 0.0124 iteration 18
AICc -82.9776 logPost -13740.2 logLik 51.3740 edf 7.8883 eps 0.0431 iteration 19
AICc -83.0956 logPost -13809.0 logLik 51.4431 edf 7.8949 eps 0.0096 iteration 20
AICc -83.2026 logPost -13815.6 logLik 51.4812 edf 7.8848 eps 0.0058 iteration 21
AICc -83.2582 logPost -13816.0 logLik 51.5109 edf 7.8861 eps 0.0574 iteration 22
AICc -83.3488 logPost -13815.9 logLik 51.5729 edf 7.8970 eps 0.0084 iteration 23
AICc -83.4298 logPost -13815.7 logLik 51.5929 edf 7.8836 eps 0.0035 iteration 24
AICc -83.4578 logPost -13815.6 logLik 51.6069 edf 7.8837 eps 0.0029 iteration 25
AICc -83.4771 logPost -13815.4 logLik 51.6166 edf 7.8837 eps 0.0025 iteration 26
AICc -83.4921 logPost -13815.3 logLik 51.6242 edf 7.8837 eps 0.0042 iteration 27
AICc -83.5087 logPost -13815.3 logLik 51.6291 edf 7.8815 eps 0.0021 iteration 28
AICc -83.5136 logPost -13815.2 logLik 51.6323 edf 7.8820 eps 0.0016 iteration 29
AICc -83.5176 logPost -13815.1 logLik 51.6343 edf 7.8820 eps 0.0013 iteration 30
AICc -83.5199 logPost -13815.1 logLik 51.6355 edf 7.8820 eps 0.0011 iteration 31
AICc -83.5219 logPost -13815.1 logLik 51.6365 edf 7.8820 eps 0.0042 iteration 32
AICc -83.5216 logPost -13815.0 logLik 51.6382 edf 7.8832 eps 0.0022 iteration 33
AICc -83.5300 logPost -13815.0 logLik 51.6383 edf 7.8806 eps 0.0009 iteration 34
AICc -83.5294 logPost -13815.0 logLik 51.6384 edf 7.8808 eps 0.0003 iteration 35
AICc -83.5293 logPost -13815.0 logLik 51.6384 edf 7.8809 eps 0.0000 iteration 36
AICc -83.5293 logPost -13815.0 logLik 51.6384 edf 7.8809 eps 0.0000 iteration 37
AICc -83.5293 logPost -13815.0 logLik 51.6384 edf 7.8809 eps 0.0000 iteration 38
AICc -83.5293 logPost -13815.0 logLik 51.6384 edf 7.8809 eps 0.0000 iteration 39
AICc -83.5293 logPost -13815.0 logLik 51.6384 edf 7.8809 eps 0.0000 iteration 39
elapsed time: 1.63sec
Starting the sampler...
| | 0% 2.18sec
|* | 5% 1.94sec 0.10sec
|** | 10% 1.70sec 0.19sec
|*** | 15% 1.55sec 0.27sec
|**** | 20% 1.41sec 0.35sec
|***** | 25% 1.32sec 0.44sec
|****** | 30% 1.23sec 0.53sec
|******* | 35% 1.16sec 0.62sec
|******** | 40% 1.08sec 0.72sec
|********* | 45% 1.01sec 0.83sec
|********** | 50% 0.92sec 0.92sec
|*********** | 55% 0.82sec 1.00sec
|************ | 60% 0.73sec 1.10sec
|************* | 65% 0.65sec 1.20sec
|************** | 70% 0.55sec 1.29sec
|*************** | 75% 0.46sec 1.38sec
|**************** | 80% 0.37sec 1.47sec
|***************** | 85% 0.28sec 1.56sec
|****************** | 90% 0.18sec 1.65sec
|******************* | 95% 0.09sec 1.75sec
|********************| 100% 0.00sec 1.85sec## Bayesian model shrinks the effects compared to ML
plot(accuracy ~ iq, data = ReadingSkills, pch = 19, col = dyslexia)
nd <- data.frame(
iq = rep(-19:20/10, 2),
dyslexia = factor(rep(c("no", "yes"), each = 40), levels = c("no", "yes"))
)
nd$betareg <- predict(rs1, newdata = nd, type = "response")
nd$bamlss <- predict(rs2, newdata = nd, type = "parameter", model = "mu")
lines(betareg ~ iq, data = nd, subset = dyslexia == "no", col = 1, lwd = 2, lty = 1)
lines(betareg ~ iq, data = nd, subset = dyslexia == "yes", col = 2, lwd = 2, lty = 1)
lines(bamlss ~ iq, data = nd, subset = dyslexia == "no", col = 1, lwd = 2, lty = 2)
lines(bamlss ~ iq, data = nd, subset = dyslexia == "yes", col = 2, lwd = 2, lty = 2)
legend("topleft", c("Dyslexia: no", "Dyslexia: yes", "betareg", "bamlss"),
lty = c(0, 0, 1, 2), pch = c(19, 19, NA, NA), col = c(1, 2, 1, 1), bty = "n")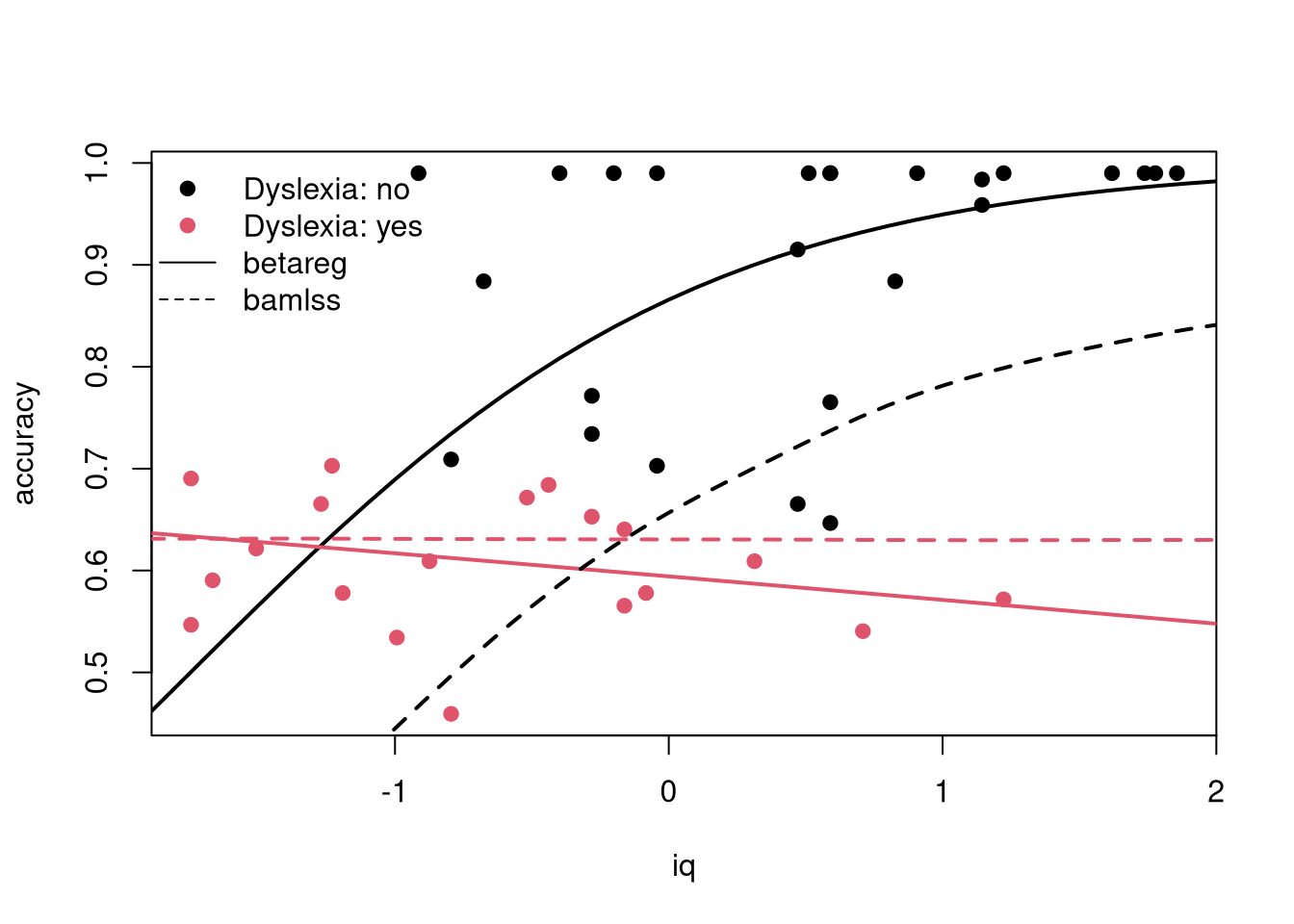
## IGNORE_RDIFF_END
## xbetax_family(): requires more time due to Gaussian quadrature
## for gamlss2: install.packages("gamlss2", repos = "https://gamlss-dev.R-universe.dev")