S3 Methods for a Reliagram (Extended Reliability Diagram)
plot.reliagram.RdGeneric plotting functions for reliability diagrams of the class "reliagram"
computed by link{reliagram}.
# S3 method for reliagram
plot(
x,
single_graph = FALSE,
minimum = 0,
confint = TRUE,
ref = TRUE,
xlim = c(0, 1),
ylim = c(0, 1),
xlab = NULL,
ylab = NULL,
main = NULL,
col = "black",
fill = adjustcolor("black", alpha.f = 0.2),
alpha_min = 0.2,
lwd = 2,
pch = 19,
lty = 1,
type = NULL,
add_hist = TRUE,
add_info = TRUE,
add_rug = TRUE,
add_min = TRUE,
axes = TRUE,
box = TRUE,
...
)
# S3 method for reliagram
lines(
x,
minimum = 0,
confint = FALSE,
ref = FALSE,
col = "black",
fill = adjustcolor("black", alpha.f = 0.2),
alpha_min = 0.2,
lwd = 2,
pch = 19,
lty = 1,
type = "b",
...
)
# S3 method for reliagram
autoplot(
object,
single_graph = FALSE,
minimum = 0,
confint = TRUE,
ref = TRUE,
xlim = c(0, 1),
ylim = c(0, 1),
xlab = NULL,
ylab = NULL,
main = NULL,
colour = "black",
fill = adjustcolor("black", alpha.f = 0.2),
alpha_min = 0.2,
size = 1,
shape = 19,
linetype = 1,
type = NULL,
add_hist = TRUE,
add_info = TRUE,
add_rug = TRUE,
add_min = TRUE,
legend = FALSE,
...
)Arguments
- single_graph
logical. Should all computed extended reliability diagrams be plotted in a single graph?
- minimum, ref, xlim, ylim, col, fill, alpha_min, lwd, pch, lty, type, add_hist, add_info, add_rug, add_min, axes, box
additional graphical parameters for base plots, whereby
xis a object of classreliagram.- confint
logical. Should confident intervals be calculated and drawn?
- xlab, ylab, main
graphical parameters.
- ...
further graphical parameters.
- object, x
an object of class
reliagram.- colour, size, shape, linetype, legend
graphical parameters passed for
ggplot2style plots, wherebyobjectis a object of classreliagram.
Details
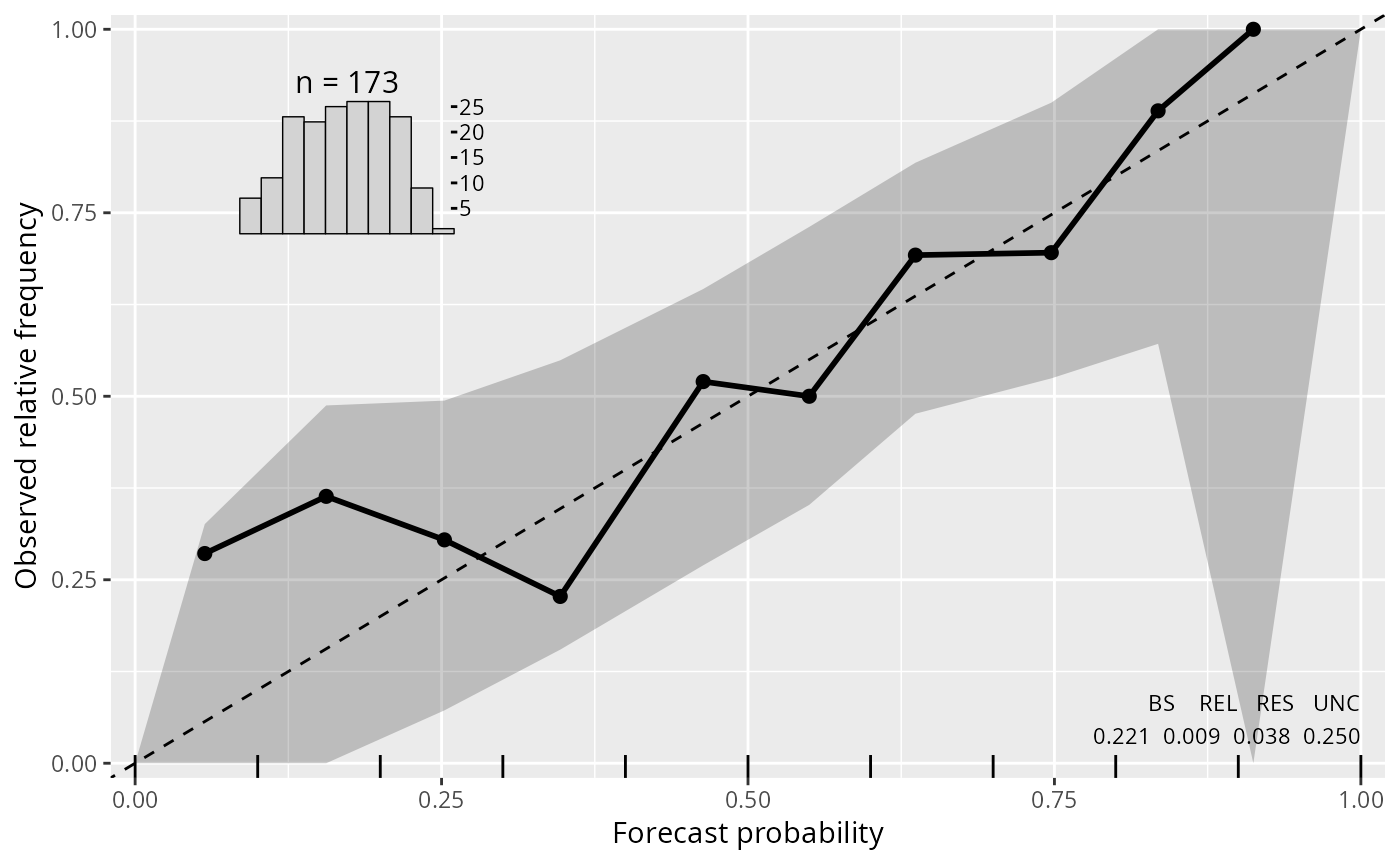
Reliagrams evaluate if a probability model is calibrated (reliable) by first partitioning the forecast probability for a binary event into a certain number of bins and then plotting (within each bin) the averaged forecast probability against the observered/empirical relative frequency.
For continous probability forecasts, reliability diagrams can be plotted either for a pre-specified threshold or for a specific quantile probability of the response values.
Reliagrams can be rendered as ggplot2 or base R graphics by using
the generics autoplot or plot.
For a single base R graphically panel, points adds an additional
reliagram.
References
Wilks DS (2011) Statistical Methods in the Atmospheric Sciences, 3rd ed., Academic Press, 704 pp.
See also
link{reliagram}, procast
Examples
## speed and stopping distances of cars
m1_lm <- lm(dist ~ speed, data = cars)
## compute and plot reliagram
reliagram(m1_lm)
 ## customize colors
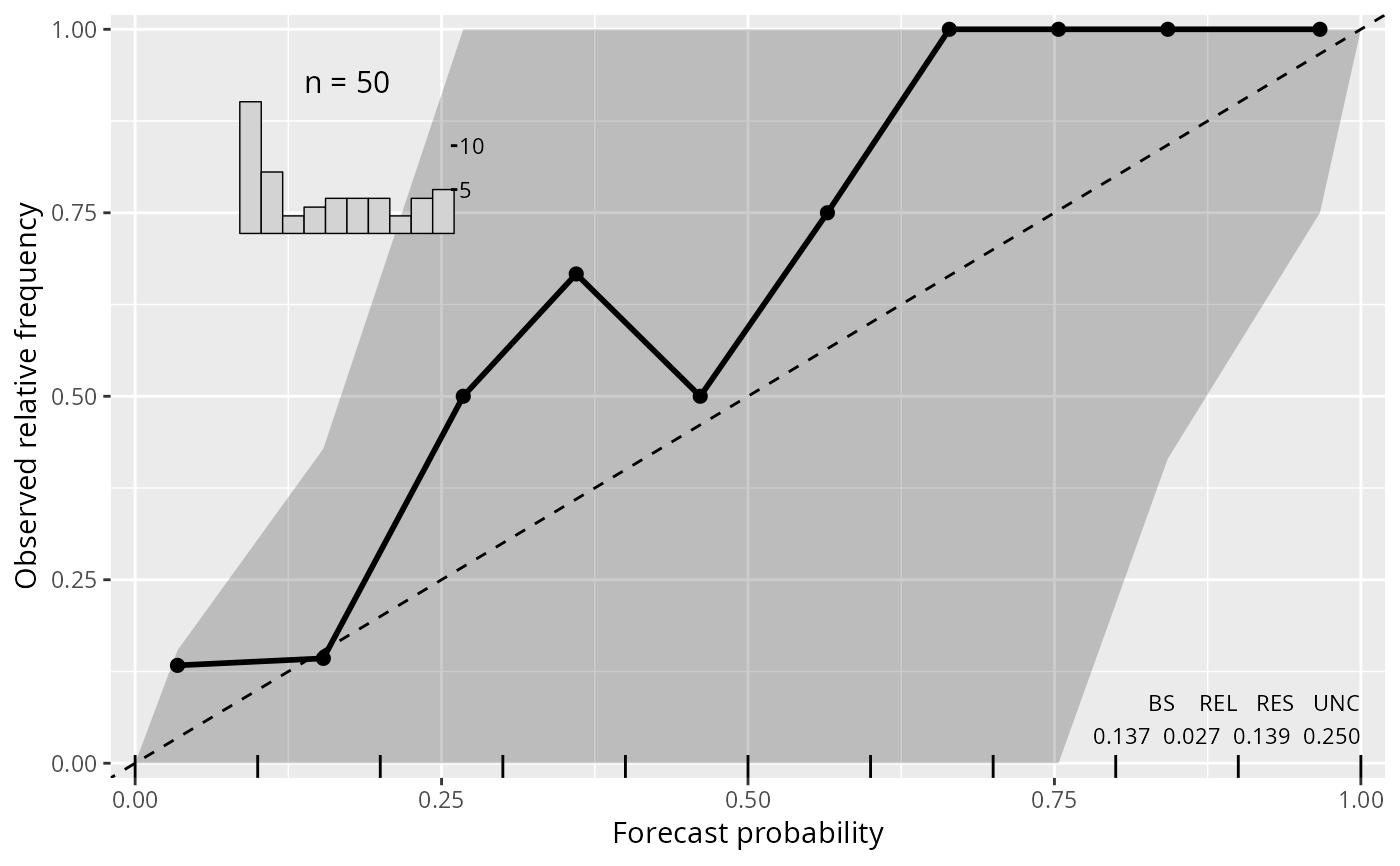
reliagram(m1_lm, ref = "blue", lty = 2, pch = 20)
## customize colors
reliagram(m1_lm, ref = "blue", lty = 2, pch = 20)
 ## add separate model
if (require("crch", quietly = TRUE)) {
m1_crch <- crch(dist ~ speed | speed, data = cars)
lines(reliagram(m1_crch, plot = FALSE), col = 2, lty = 2, confint = 2)
}
#> Error in polygon(na.omit(c(ifelse(extend_left, 0, NA), d[min_idx, "x"], ifelse(extend_right, 1, NA), rev(d[min_idx, "x"]), ifelse(extend_left, 0, NA))), na.omit(c(ifelse(extend_left, 0, NA), d[min_idx, "ci_lwr"], ifelse(extend_right, 1, NA), rev(d[min_idx, "ci_upr"]), ifelse(extend_left, 0, NA))), col = set_minimum_transparency(plot_arg$confint[j], alpha_min = plot_arg$alpha_min[j]), border = NA): plot.new has not been called yet
#-------------------------------------------------------------------------------
if (require("crch")) {
## precipitation observations and forecasts for Innsbruck
data("RainIbk", package = "crch")
RainIbk <- sqrt(RainIbk)
RainIbk$ensmean <- apply(RainIbk[,grep('^rainfc',names(RainIbk))], 1, mean)
RainIbk$enssd <- apply(RainIbk[,grep('^rainfc',names(RainIbk))], 1, sd)
RainIbk <- subset(RainIbk, enssd > 0)
## linear model w/ constant variance estimation
m2_lm <- lm(rain ~ ensmean, data = RainIbk)
## logistic censored model
m2_crch <- crch(rain ~ ensmean | log(enssd), data = RainIbk, left = 0, dist = "logistic")
## compute reliagrams
rel2_lm <- reliagram(m2_lm, plot = FALSE)
rel2_crch <- reliagram(m2_crch, plot = FALSE)
## plot in single graph
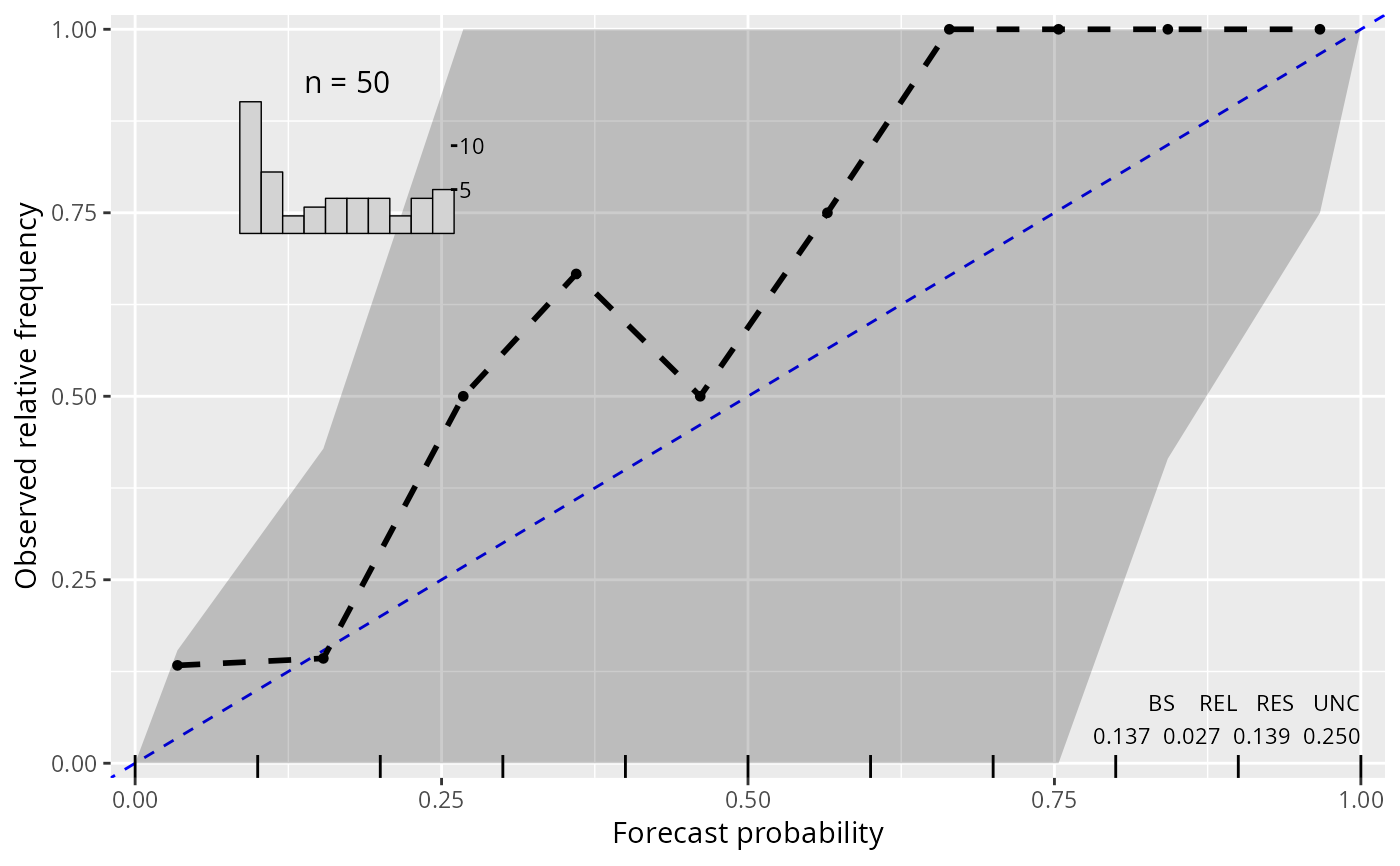
plot(c(rel2_lm, rel2_crch), col = c(1, 2), confint = c(1, 2), ref = 3, single_graph = TRUE)
}
#-------------------------------------------------------------------------------
## determinants for male satellites to nesting horseshoe crabs
data("CrabSatellites", package = "countreg")
## linear poisson model
m3_pois <- glm(satellites ~ width + color, data = CrabSatellites, family = poisson)
## compute and plot reliagram as "ggplot2" graphic
reliagram(m3_pois, plot = "ggplot2")
## add separate model
if (require("crch", quietly = TRUE)) {
m1_crch <- crch(dist ~ speed | speed, data = cars)
lines(reliagram(m1_crch, plot = FALSE), col = 2, lty = 2, confint = 2)
}
#> Error in polygon(na.omit(c(ifelse(extend_left, 0, NA), d[min_idx, "x"], ifelse(extend_right, 1, NA), rev(d[min_idx, "x"]), ifelse(extend_left, 0, NA))), na.omit(c(ifelse(extend_left, 0, NA), d[min_idx, "ci_lwr"], ifelse(extend_right, 1, NA), rev(d[min_idx, "ci_upr"]), ifelse(extend_left, 0, NA))), col = set_minimum_transparency(plot_arg$confint[j], alpha_min = plot_arg$alpha_min[j]), border = NA): plot.new has not been called yet
#-------------------------------------------------------------------------------
if (require("crch")) {
## precipitation observations and forecasts for Innsbruck
data("RainIbk", package = "crch")
RainIbk <- sqrt(RainIbk)
RainIbk$ensmean <- apply(RainIbk[,grep('^rainfc',names(RainIbk))], 1, mean)
RainIbk$enssd <- apply(RainIbk[,grep('^rainfc',names(RainIbk))], 1, sd)
RainIbk <- subset(RainIbk, enssd > 0)
## linear model w/ constant variance estimation
m2_lm <- lm(rain ~ ensmean, data = RainIbk)
## logistic censored model
m2_crch <- crch(rain ~ ensmean | log(enssd), data = RainIbk, left = 0, dist = "logistic")
## compute reliagrams
rel2_lm <- reliagram(m2_lm, plot = FALSE)
rel2_crch <- reliagram(m2_crch, plot = FALSE)
## plot in single graph
plot(c(rel2_lm, rel2_crch), col = c(1, 2), confint = c(1, 2), ref = 3, single_graph = TRUE)
}
#-------------------------------------------------------------------------------
## determinants for male satellites to nesting horseshoe crabs
data("CrabSatellites", package = "countreg")
## linear poisson model
m3_pois <- glm(satellites ~ width + color, data = CrabSatellites, family = poisson)
## compute and plot reliagram as "ggplot2" graphic
reliagram(m3_pois, plot = "ggplot2")