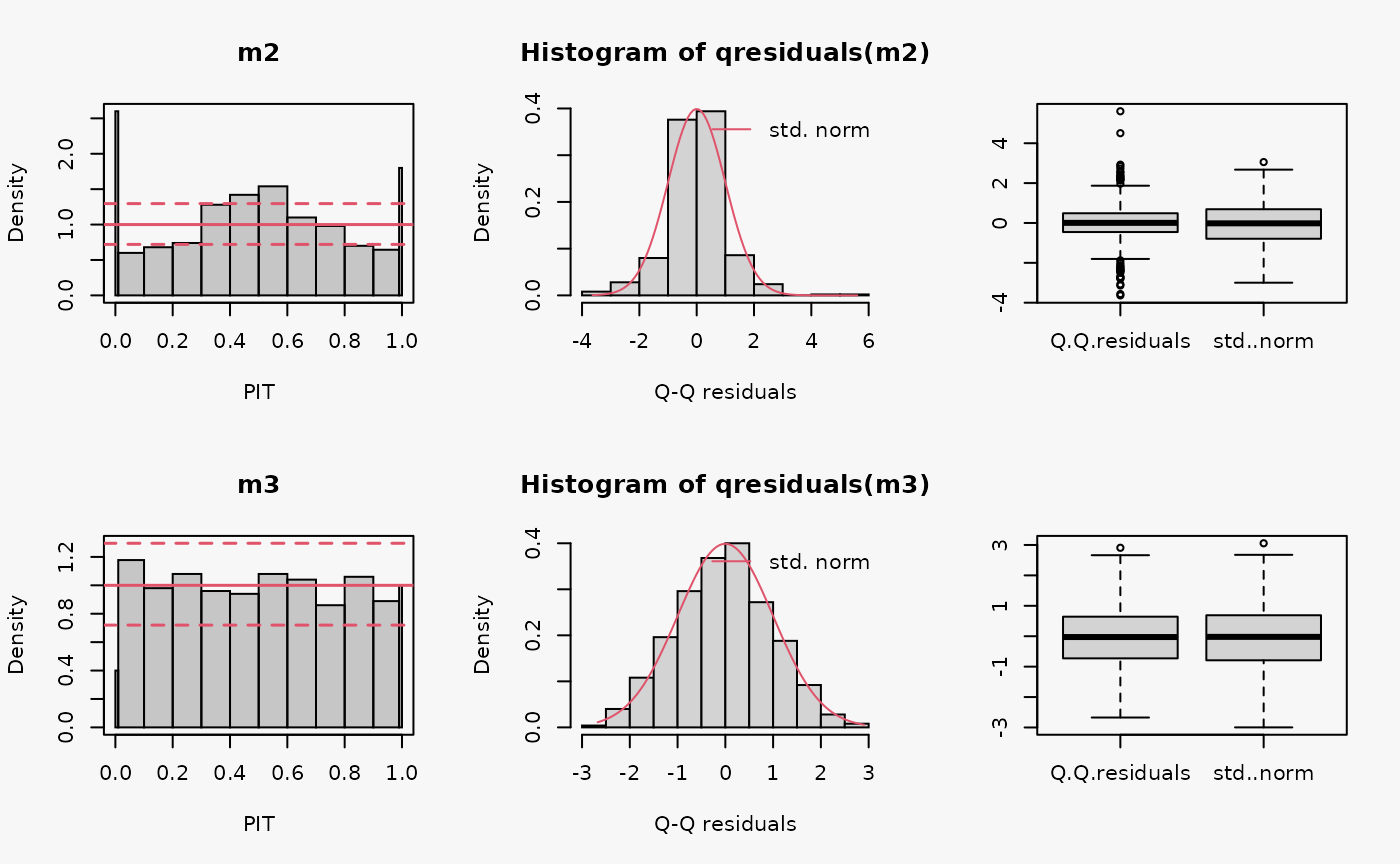
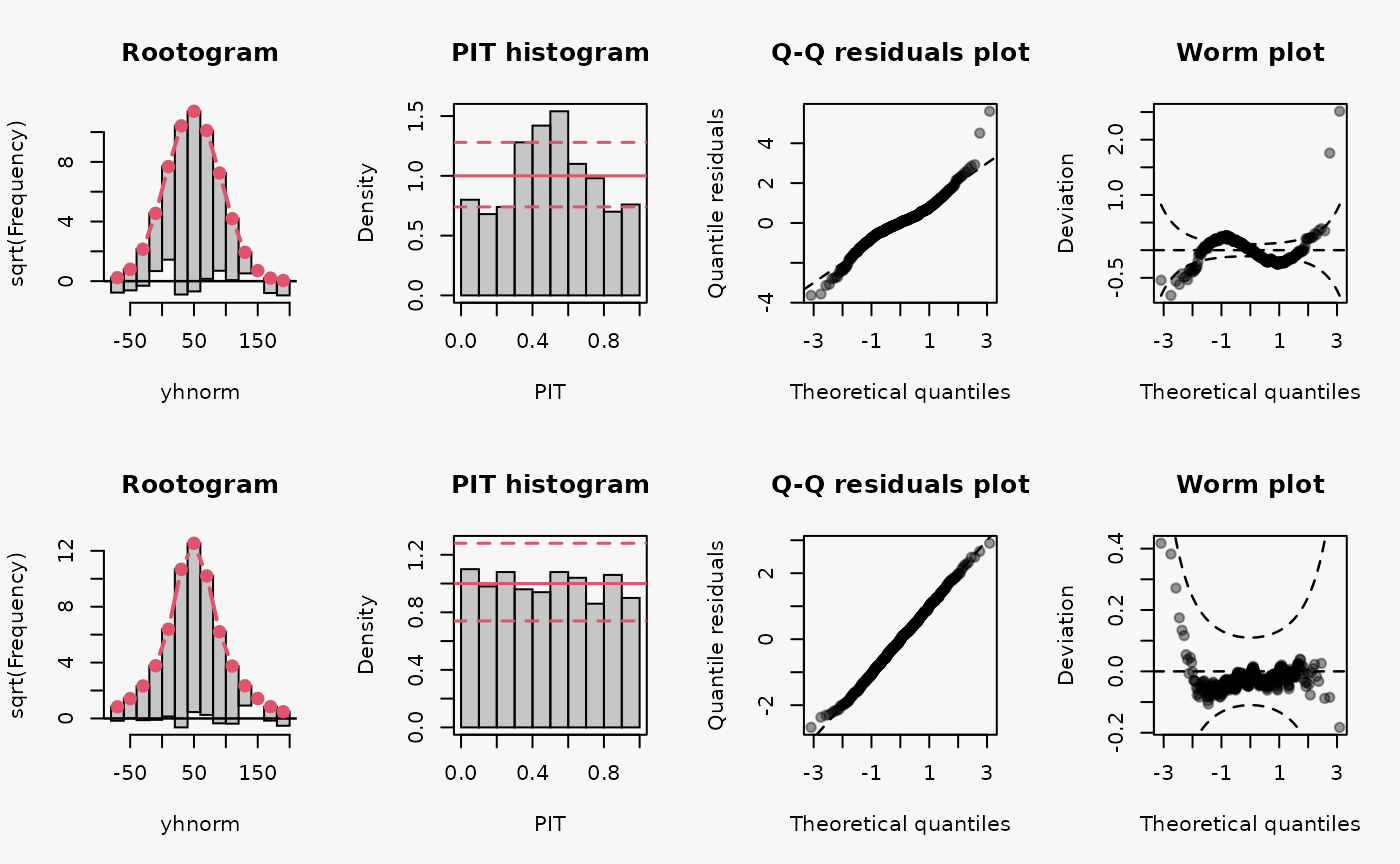
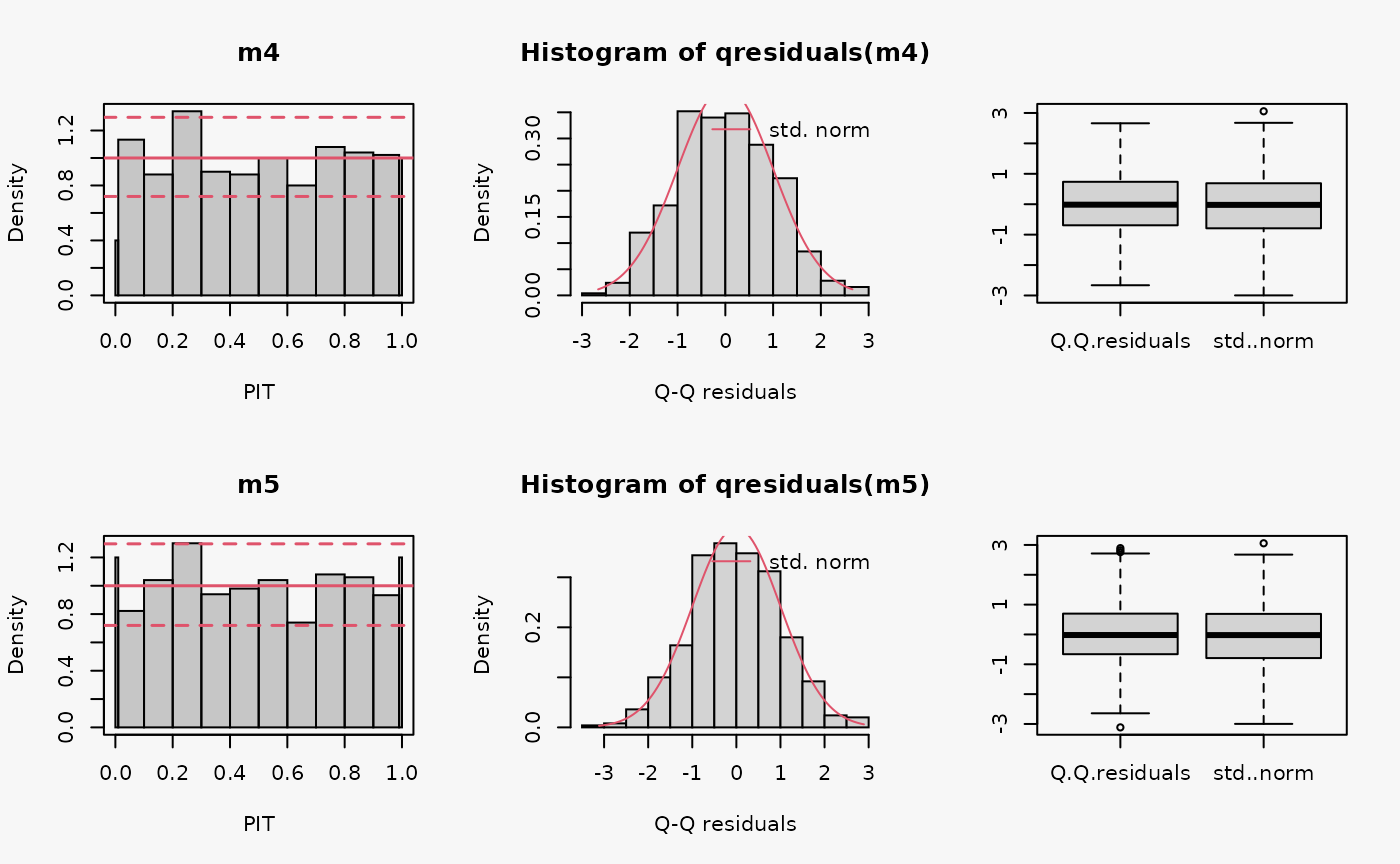
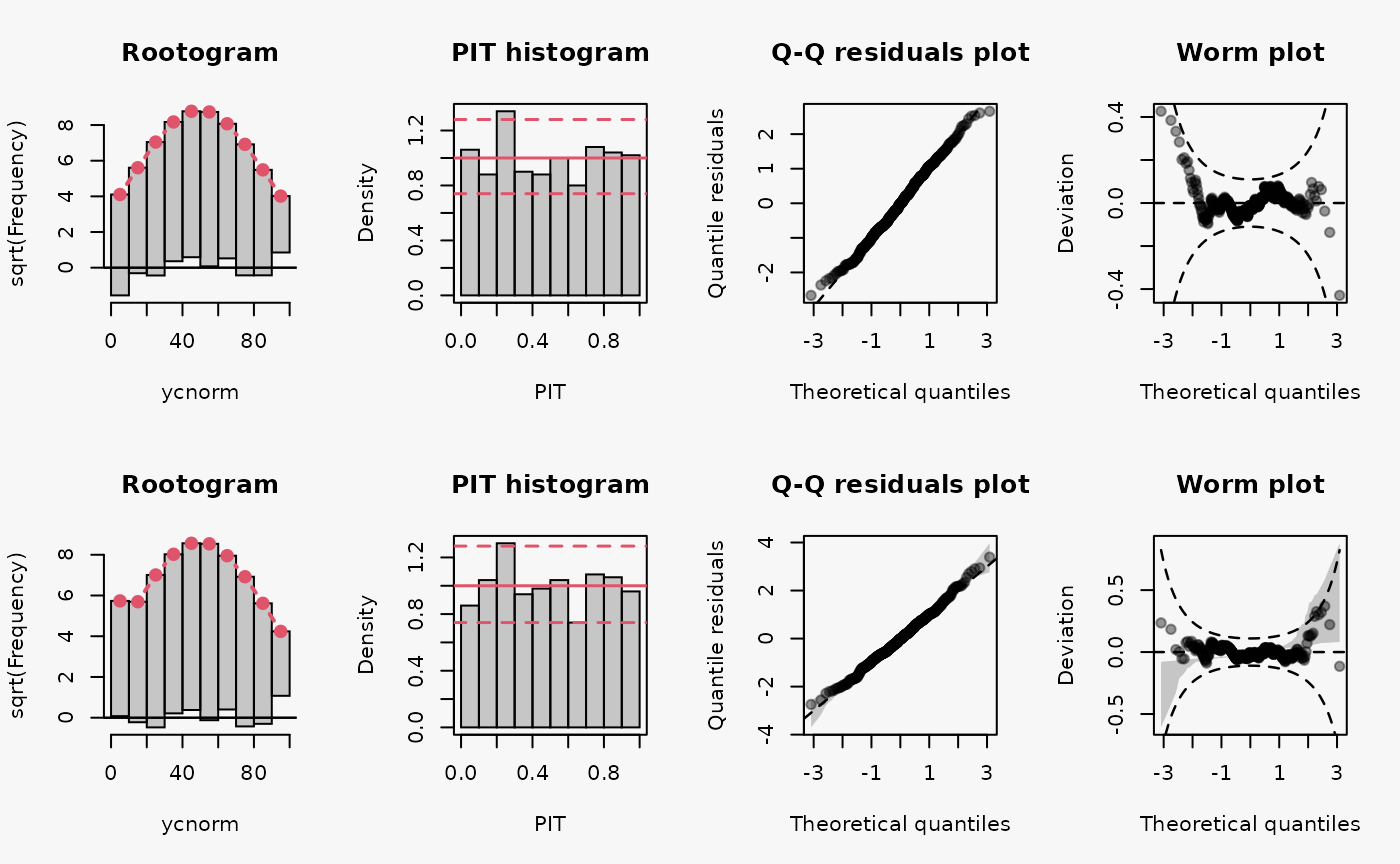
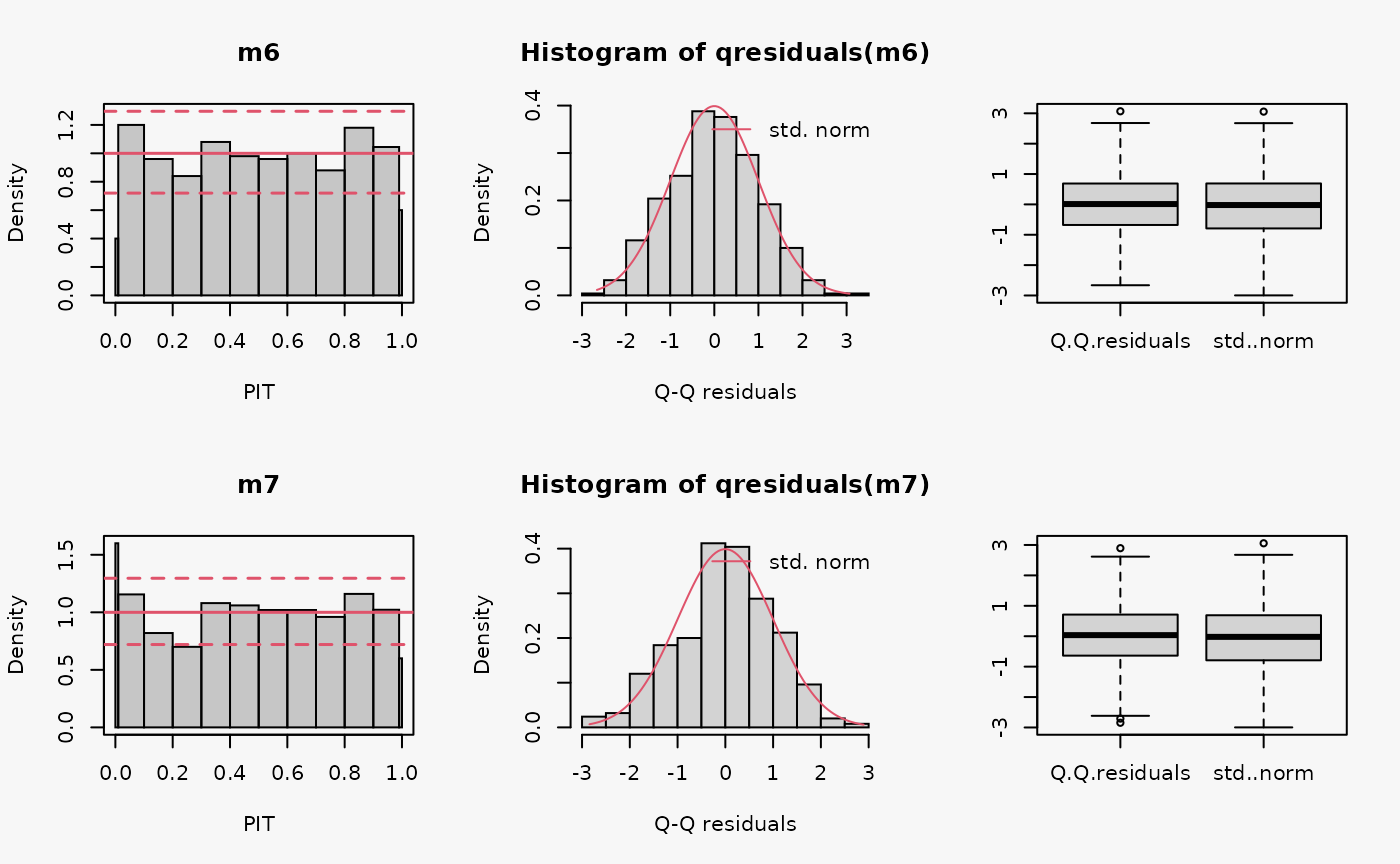
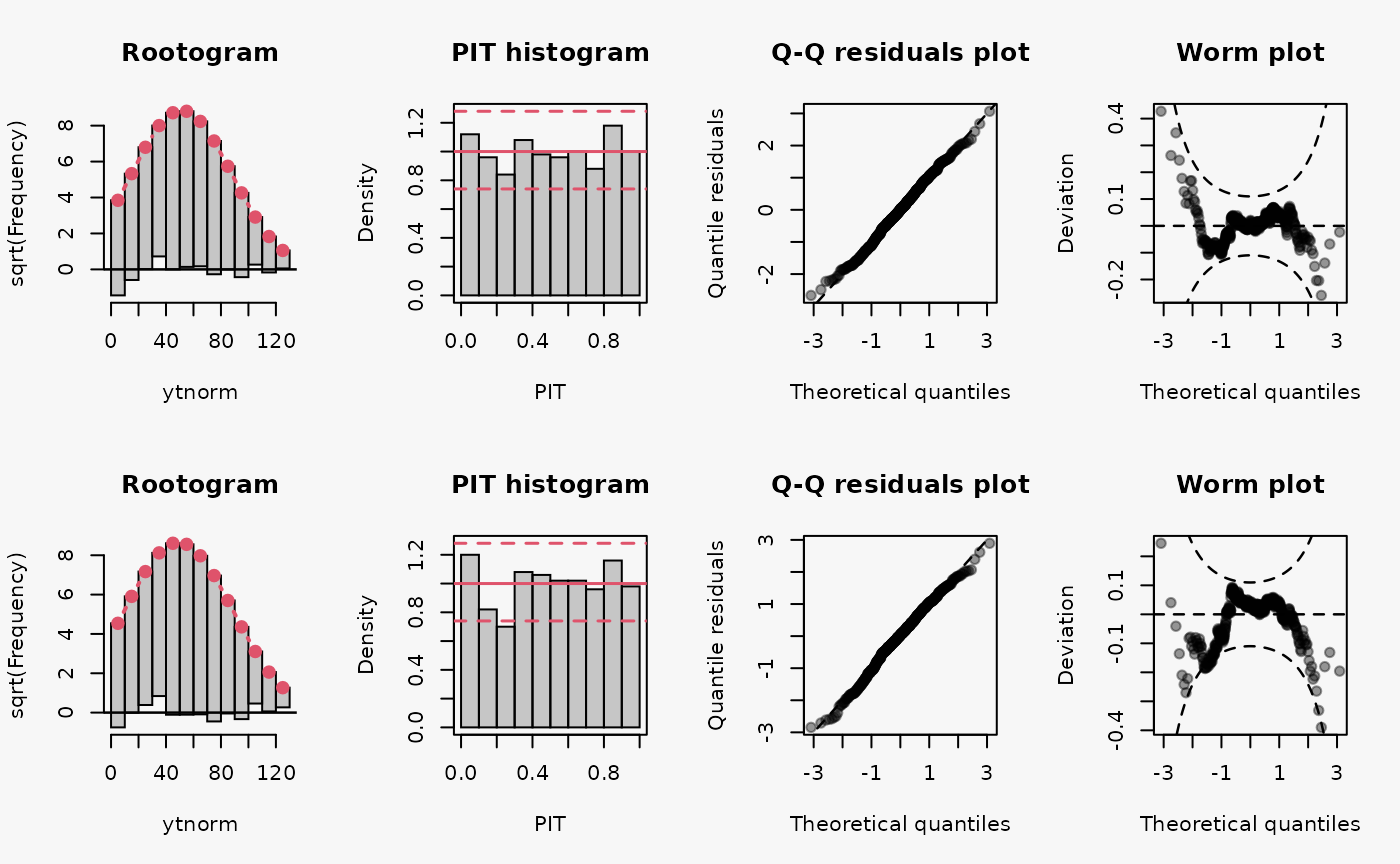
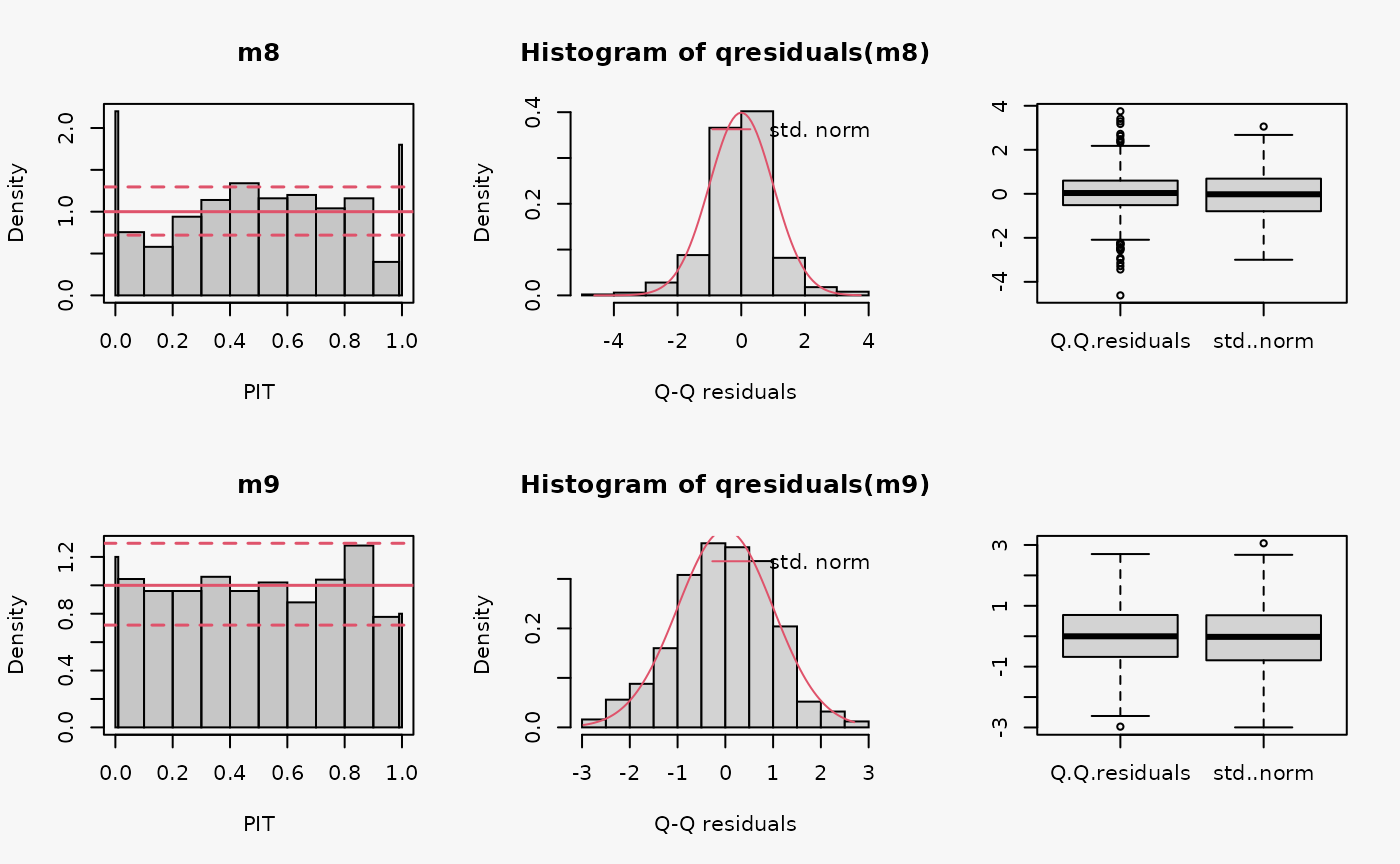
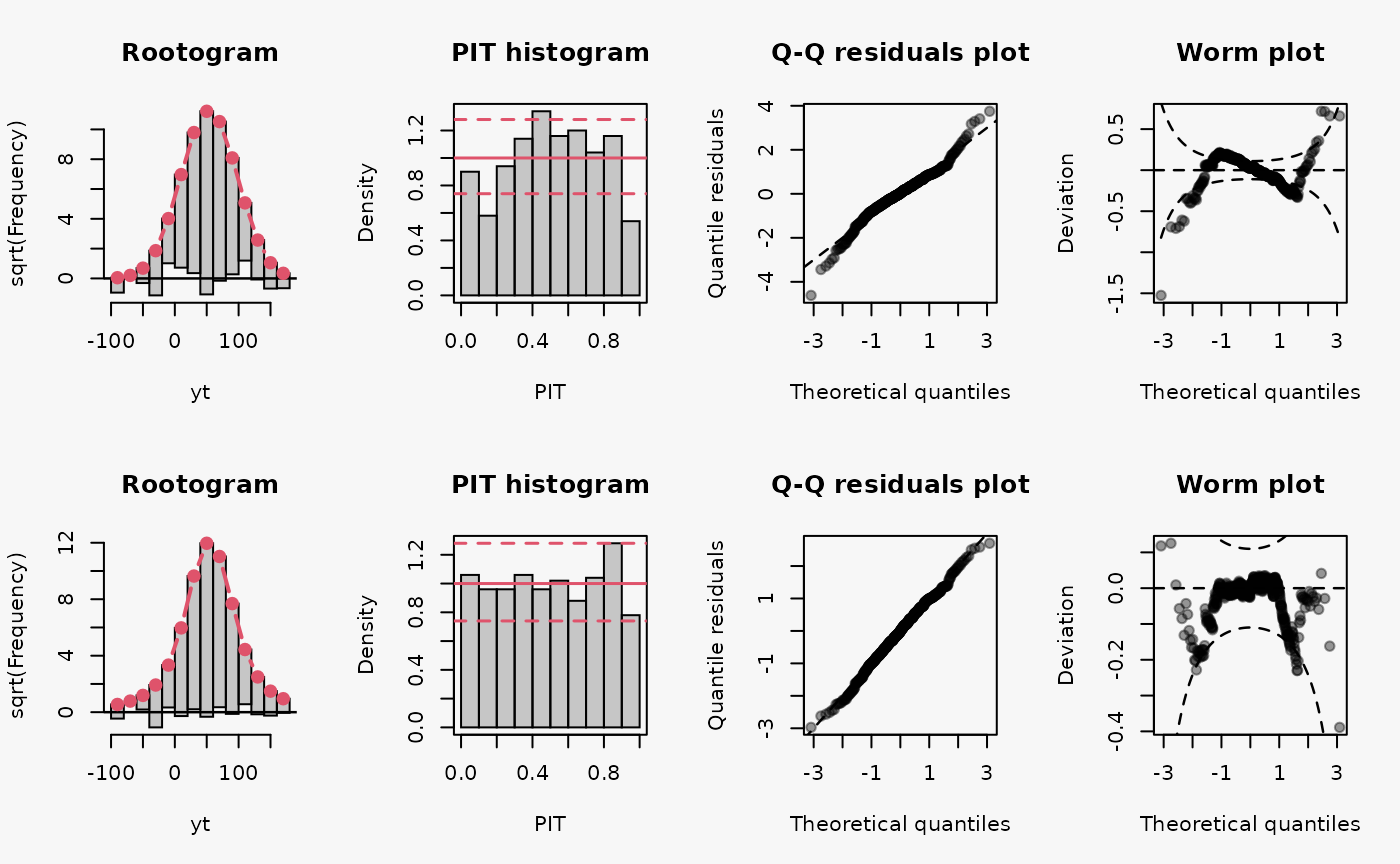
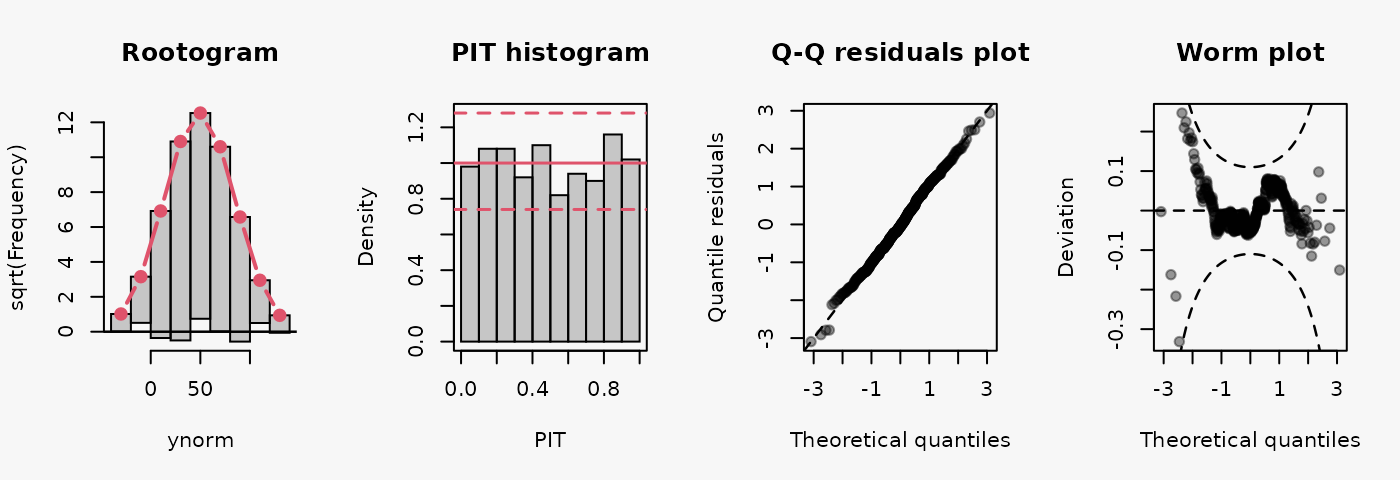
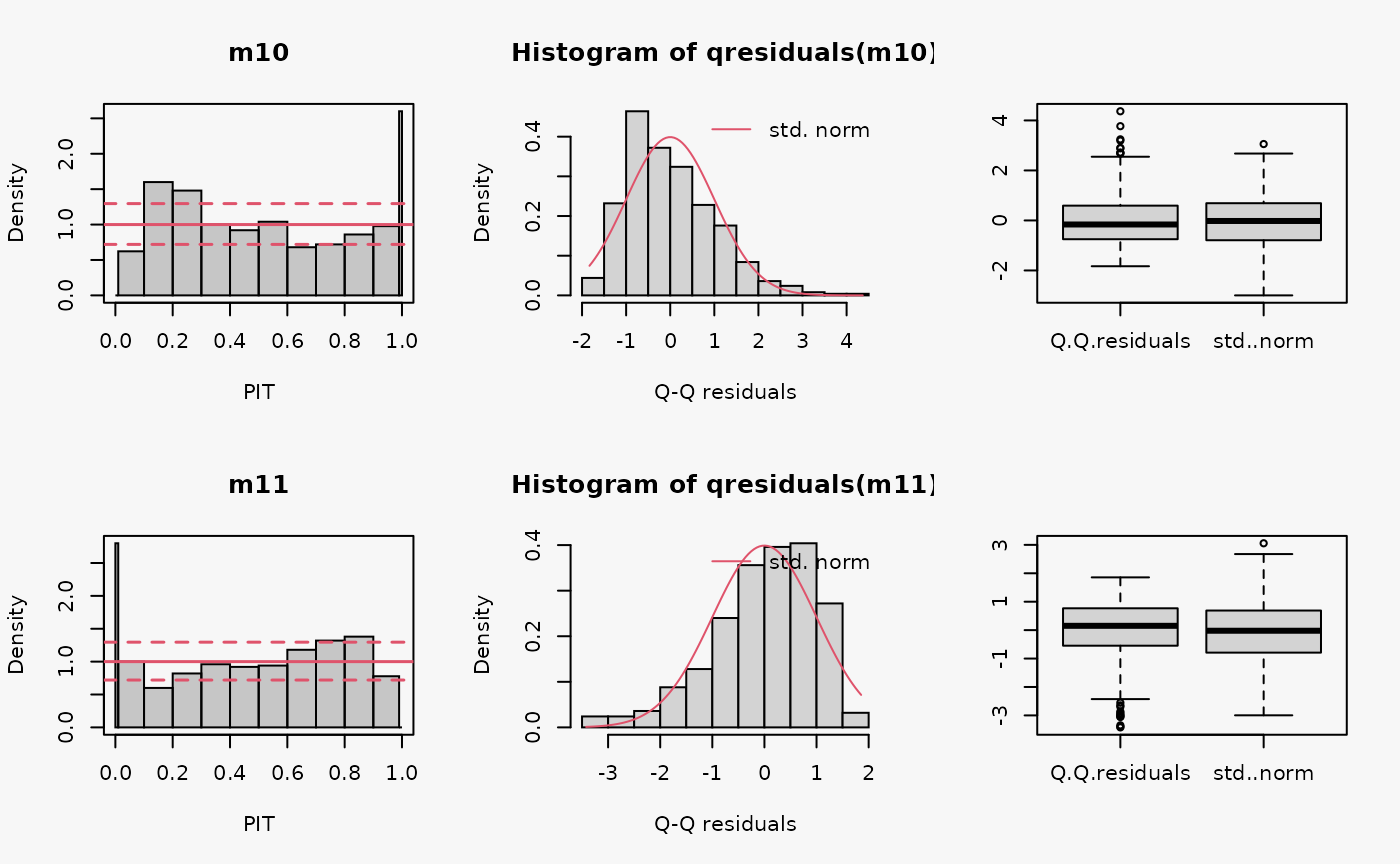
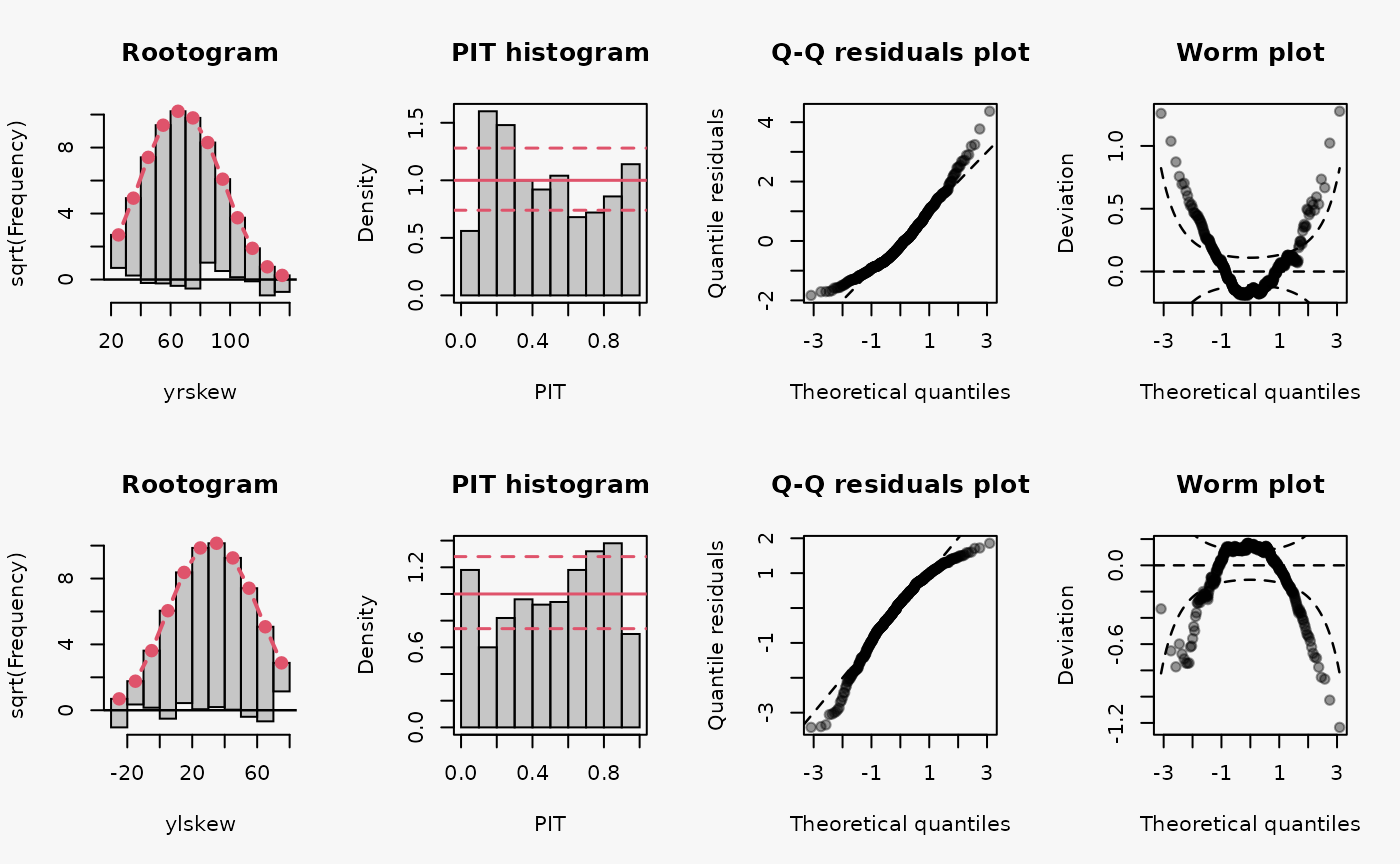
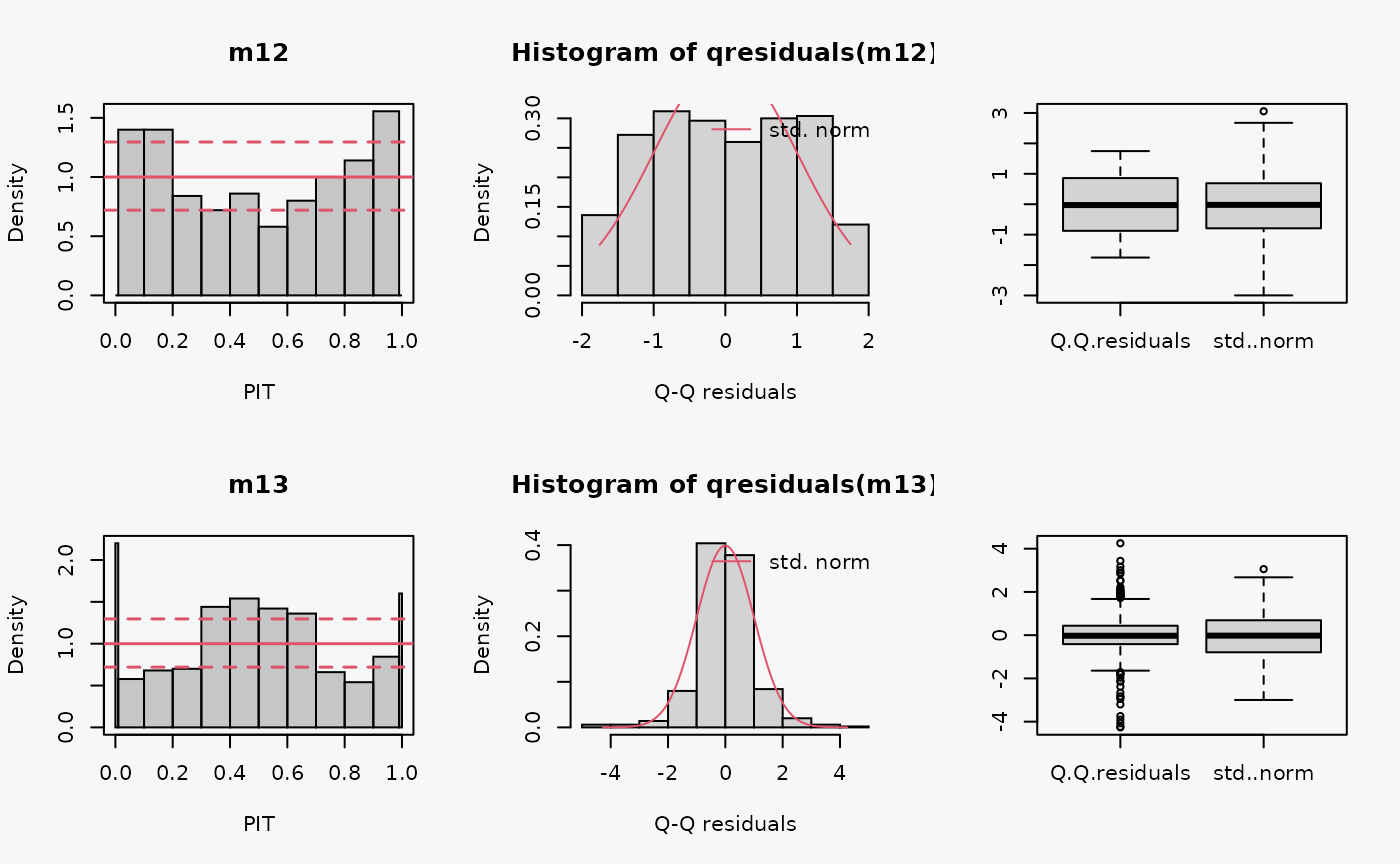
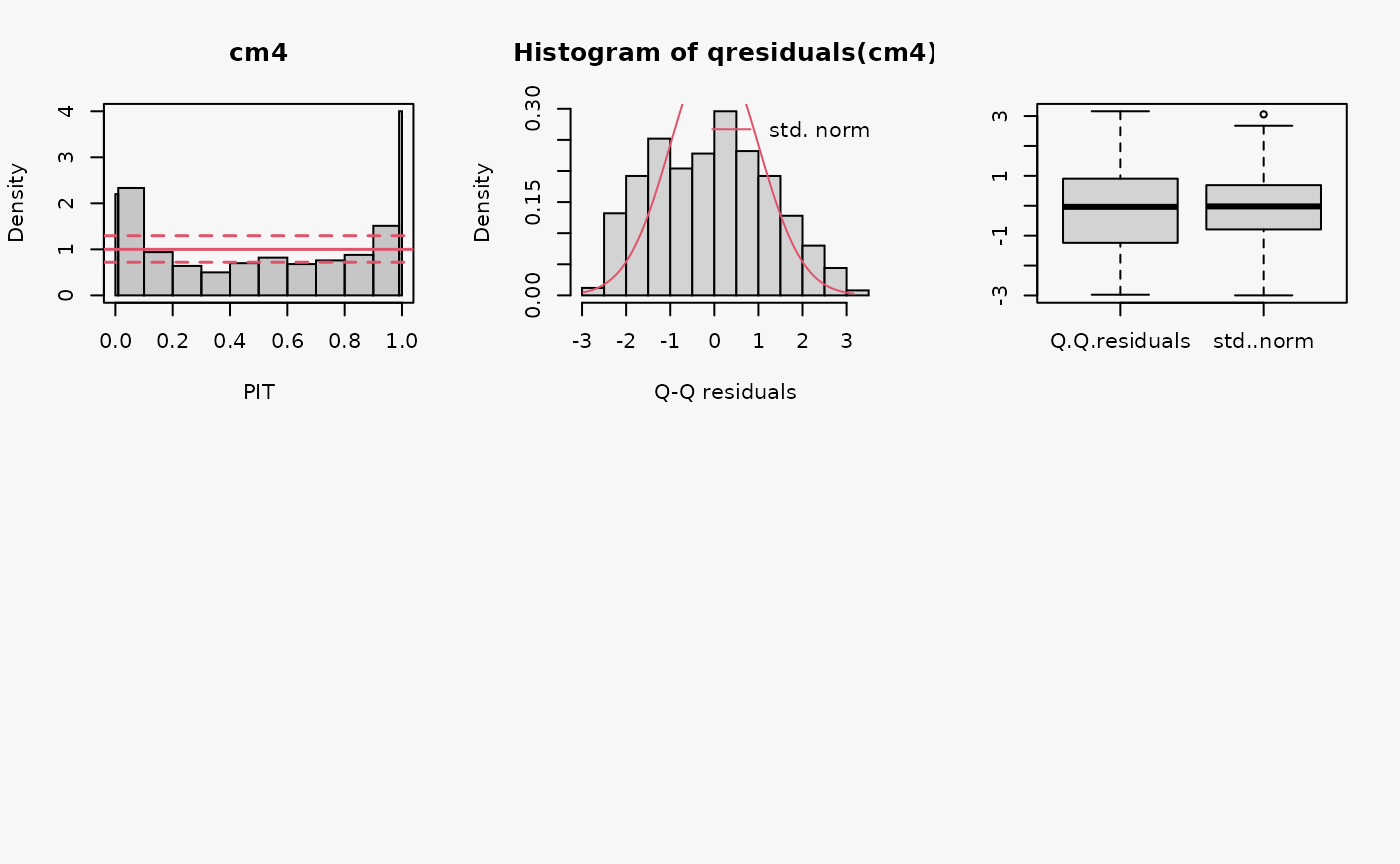
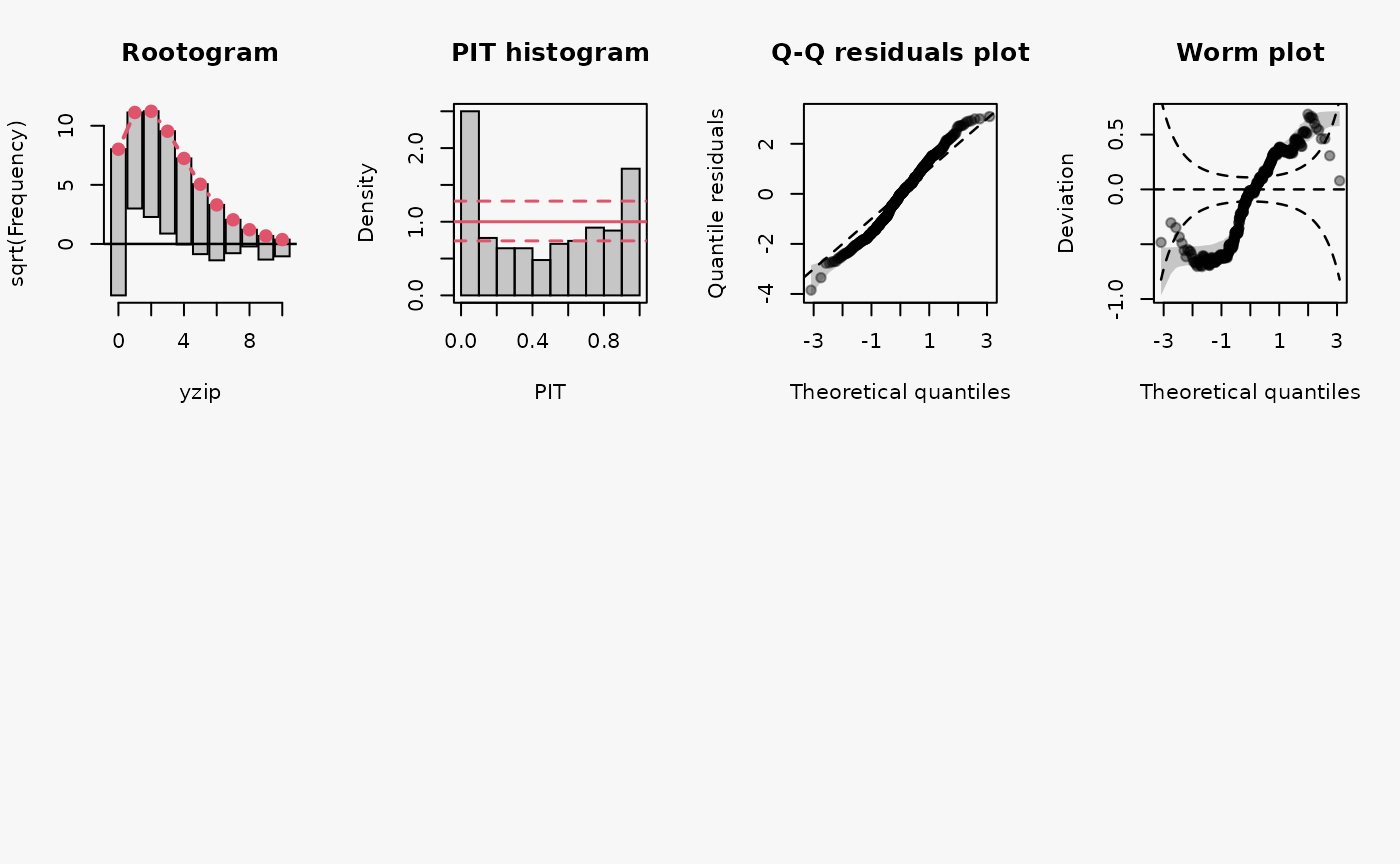
set.seed(0)
## regressors
d <- data.frame(
x = runif(500, -1, 1),
z = runif(500, -1, 1)
)
## parameters
d <- transform(d,
lambda = exp(1 + 0.5 * x),
theta = 2,
mu = 50 + 22 * x,
sigma = 22,
sigmaz = exp(3 + 1 * z)
)
## responses
d <- transform(d,
ynorm = rnorm(500, mean = mu, sd = sigma),
yhnorm = rnorm(500, mean = mu, sd = sigmaz),
ytnorm = crch::rtnorm(500, mean = mu, sd = sigma, left = 0),
ycnorm = crch::rcnorm(500, mean = mu, sd = sigma, left = 0, right = 100),
yt = mu + sigma * rt(500, df = 4),
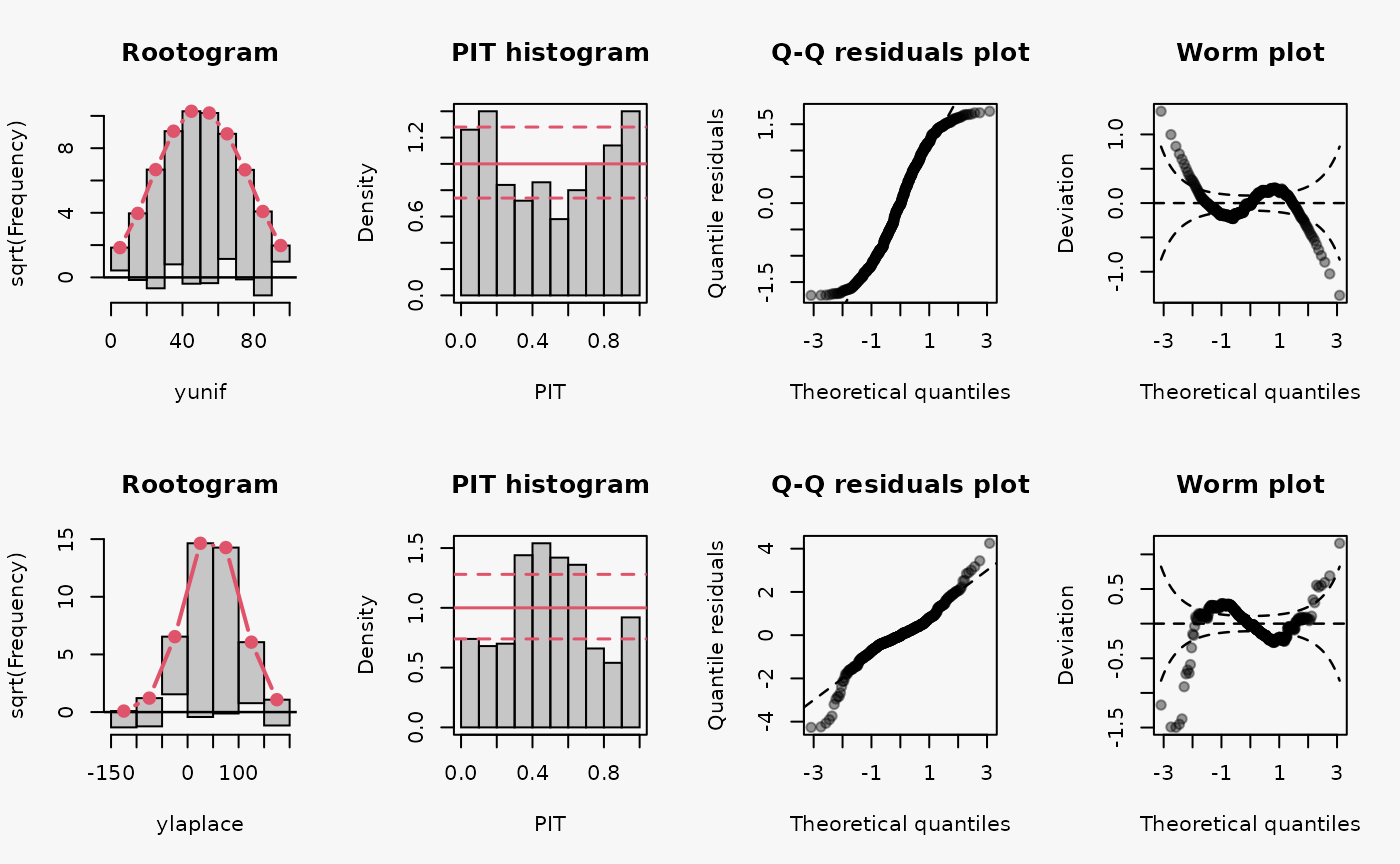
ylaplace = mu + rmutil::rlaplace(500, s = sigma),
yrskew = sn::rsn(500, xi = mu, omega = sigma, alpha = 5, tau = 0),
ylskew = sn::rsn(500, xi = mu, omega = sigma, alpha = -5, tau = 0),
yunif = mu + sigma * runif(500, min = -1, max = 1),
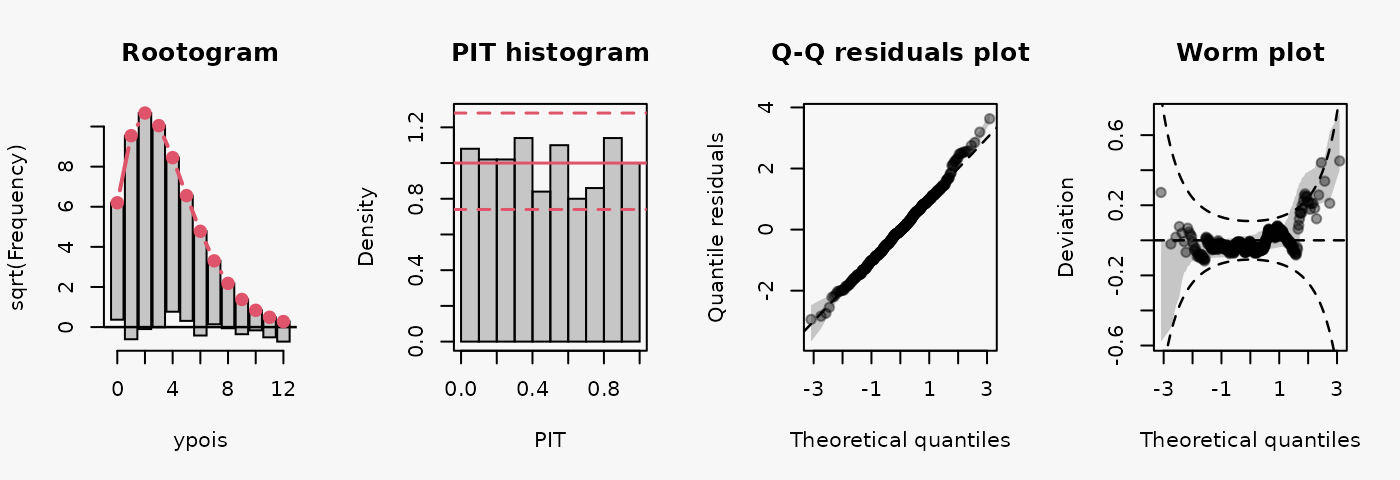
ypois = rpois(500, lambda = lambda),
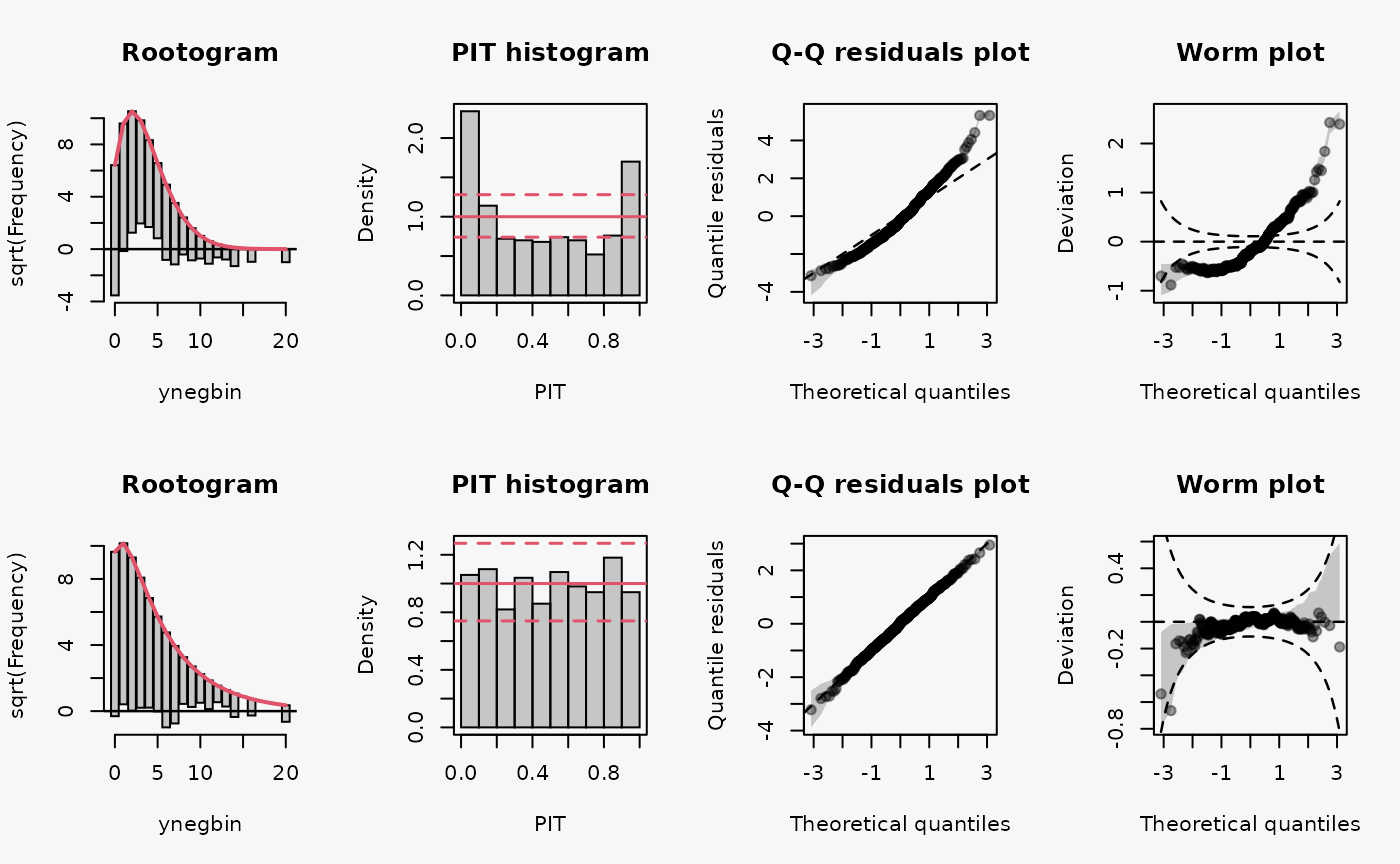
ynegbin = rnbinom(500, mu = lambda, size = theta),
yzip = ifelse(runif(500) < 0.25, 0, rpois(500, lambda = lambda))
)