geom_* and stat_* for Producing PIT Histograms with `ggplot2`
geom_rootogram.RdVarious geom_* and stat_* used within
autoplot for producing PIT histograms.
stat_rootogram(
mapping = NULL,
data = NULL,
geom = "rootogram",
position = "identity",
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE,
scale = c("sqrt", "raw"),
style = c("hanging", "standing", "suspended"),
...
)
geom_rootogram(
mapping = NULL,
data = NULL,
stat = "rootogram",
position = "identity",
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE,
scale = c("sqrt", "raw"),
style = c("hanging", "standing", "suspended"),
...
)
stat_rootogram_expected(
mapping = NULL,
data = NULL,
geom = "rootogram_expected",
position = "identity",
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE,
scale = c("sqrt", "raw"),
...
)
geom_rootogram_expected(
mapping = NULL,
data = NULL,
stat = "rootogram_expected",
position = "identity",
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE,
scale = c("sqrt", "raw"),
linestyle = c("both", "line", "point"),
...
)
GeomRootogramExpected
geom_rootogram_ref(
mapping = NULL,
data = NULL,
stat = "identity",
position = "identity",
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE,
...
)
stat_rootogram_confint(
mapping = NULL,
data = NULL,
geom = "rootogram_confint",
position = "identity",
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE,
level = 0.95,
nrep = 1000,
type = c("pointwise", "simultaneous"),
scale = c("sqrt", "raw"),
rootogram_style = c("hanging", "standing", "suspended"),
...
)
geom_rootogram_confint(
mapping = NULL,
data = NULL,
stat = "rootogram_confint",
position = "identity",
na.rm = FALSE,
show.legend = NA,
inherit.aes = TRUE,
level = 0.95,
nrep = 1000,
type = c("pointwise", "simultaneous"),
scale = c("sqrt", "raw"),
rootogram_style = c("hanging", "standing", "suspended"),
...
)Format
An object of class GeomRootogramExpected (inherits from GeomPath, Geom, ggproto, gg) of length 3.
Arguments
- mapping
Set of aesthetic mappings created by
aes(). If specified andinherit.aes = TRUE(the default), it is combined with the default mapping at the top level of the plot. You must supplymappingif there is no plot mapping.- data
The data to be displayed in this layer. There are three options:
If
NULL, the default, the data is inherited from the plot data as specified in the call toggplot().A
data.frame, or other object, will override the plot data. All objects will be fortified to produce a data frame. Seefortify()for which variables will be created.A
functionwill be called with a single argument, the plot data. The return value must be adata.frame, and will be used as the layer data. Afunctioncan be created from aformula(e.g.~ head(.x, 10)).- geom
The geometric object to use to display the data, either as a
ggprotoGeomsubclass or as a string naming the geom stripped of thegeom_prefix (e.g."point"rather than"geom_point")- position
Position adjustment, either as a string naming the adjustment (e.g.
"jitter"to useposition_jitter), or the result of a call to a position adjustment function. Use the latter if you need to change the settings of the adjustment.- na.rm
If
FALSE, the default, missing values are removed with a warning. IfTRUE, missing values are silently removed.- show.legend
logical. Should this layer be included in the legends?
NA, the default, includes if any aesthetics are mapped.FALSEnever includes, andTRUEalways includes. It can also be a named logical vector to finely select the aesthetics to display.- inherit.aes
If
FALSE, overrides the default aesthetics, rather than combining with them. This is most useful for helper functions that define both data and aesthetics and shouldn't inherit behaviour from the default plot specification, e.g.borders().- scale
character specifying whether values should be transformed to the square root scale (not checking for original scale, so maybe applied again).
- style
character specifying the syle of rootogram (see below).
- ...
Other arguments passed on to
layer(). These are often aesthetics, used to set an aesthetic to a fixed value, likecolour = "red"orsize = 3. They may also be parameters to the paired geom/stat.- stat
The statistical transformation to use on the data for this layer, either as a
ggprotoGeomsubclass or as a string naming the stat stripped of thestat_prefix (e.g."count"rather than"stat_count")- linestyle
Character string defining one of `"both"`, `"line"` or `"point"`.
- level
numeric. The confidence level required.
- nrep
numeric. The repetition number of simulation for computing the confidence intervals.
- type
character. Should
"pointwise"or"simultaneous"confidence intervals be visualized.- rootogram_style
character specifying the syle of rootogram.
Examples
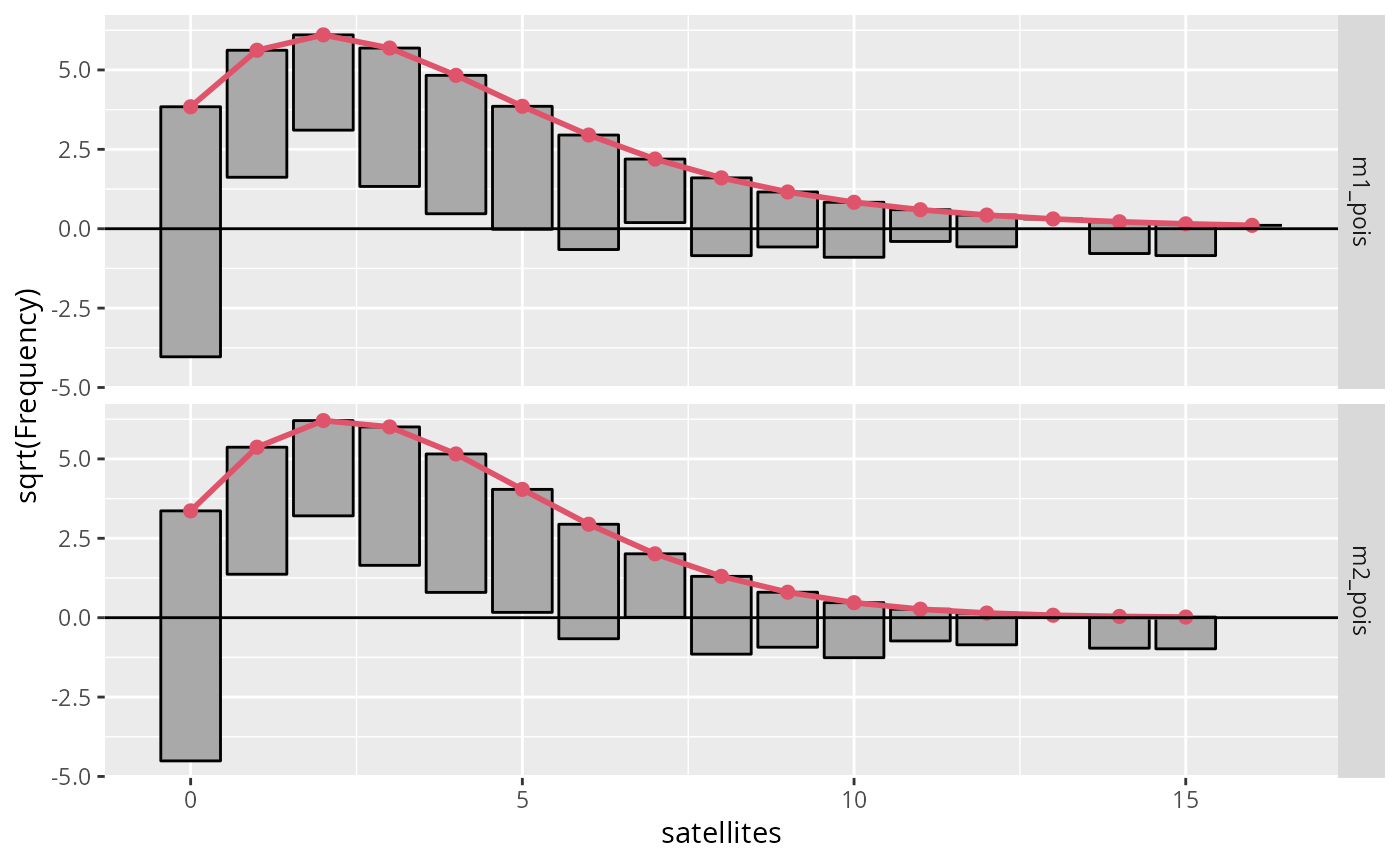
if (require("ggplot2")) {
## Fit model
data("CrabSatellites", package = "countreg")
m1_pois <- glm(satellites ~ width + color, data = CrabSatellites, family = poisson)
m2_pois <- glm(satellites ~ color, data = CrabSatellites, family = poisson)
## Compute rootogram (on raw scale)
p1 <- rootogram(m1_pois, scale = "raw", plot = FALSE)
p2 <- rootogram(m2_pois, scale = "raw", plot = FALSE)
d <- c(p1, p2)
## Get label names
main <- attr(d, "main")
main <- make.names(main, unique = TRUE)
d$group <- factor(d$group, labels = main)
## Plot rootograms w/ on default "sqrt" scale
gg1 <- ggplot(data = d) +
geom_rootogram(aes(
observed = observed, expected = expected, mid = mid,
width = width, group = group
)) +
geom_rootogram_expected(aes(expected = expected, mid = mid)) +
geom_rootogram_ref() +
facet_grid(group ~ .) +
xlab("satellites") +
ylab("sqrt(Frequency)")
gg1
}